AXON LOSS
|
Axon loss Early Chronic Myelinated Large Large & Small Large vs Small Differential fascicular Skin Schwann cell Δ Bungner bands Collagen pockets Injury patterns Wallerian degeneration Also see Regeneration Axon sprouts |
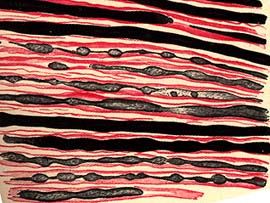
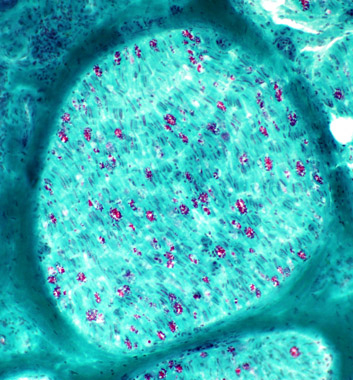 Gomori trichrome stain Myelinated Axons (Red): Loss
Degree: Moderately severe Myelin abnormal: Irregularly stained |
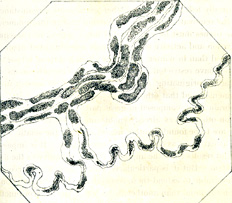
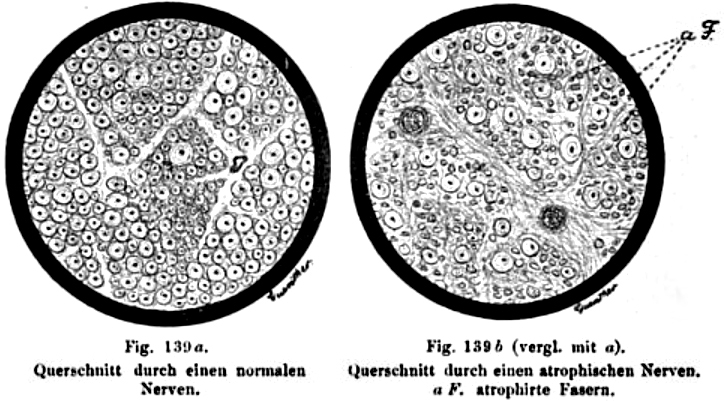 Oppenheim 1894 |
Nerve Injury
Degrees
- Neuropraxia
- Axon: Anatomically intact: No Wallerian degeneration
- Anatomy
- Especially affects: Large myelinated axons
- Pathology: Focal demyelination or dysfuntion
- Physiology: Nerve conduction block
- Clinical pattern: Motor > Sensory
- Axonotmesis
- Axons: Discontinuous Wallerian degeneration
- Epineurium: Continuous
- Neurotmesis
- Complete nerve disconnection
Electrophysiology: Changes after nerve transection
- CMAP
- Normal for 2 to 3 days
- Reduced amplitude: Reaches nadir at 7 to 10 days
- SNAP
- Amplitude: Reaches nadir at day 10 or 11
- EMG
- Recruitment reduced: Immediately after transection
- Fibrillation potentials &
Positive waves: Onset
- Typical: 14 days
- May be shorter with lesion close to muscle
Wallerian degeneration
- Motor axons: Onset 3 to 5 days
- Sensory axons: Onset 6 to 10 days
Axon Loss: Small > Large
Myelinated axons: Loss
|
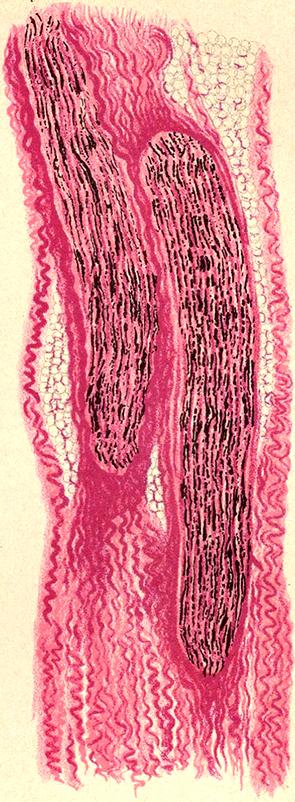
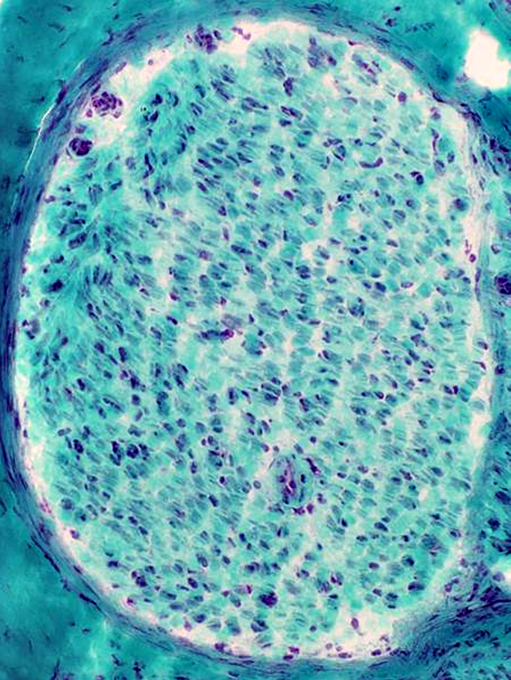
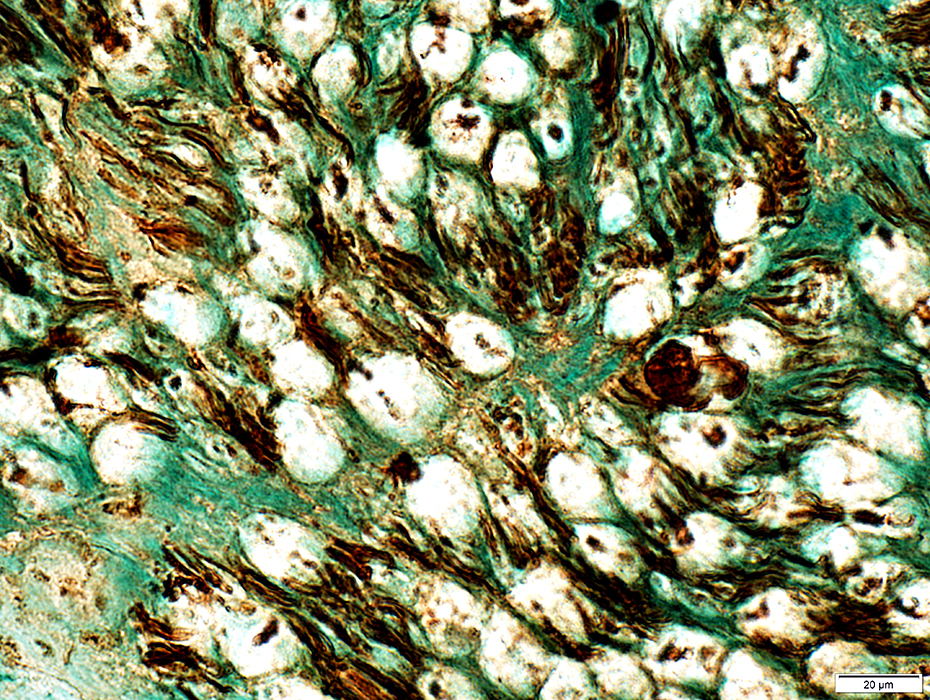
Myelinated Axon loss: Moderate 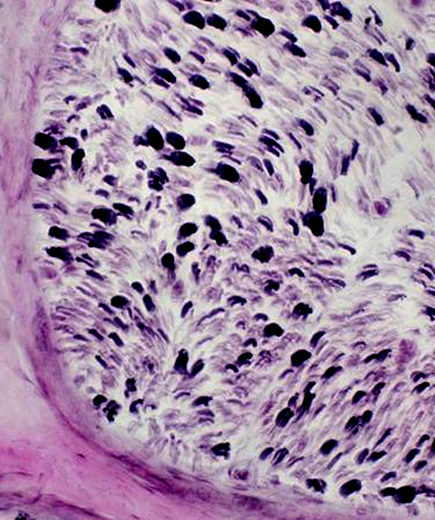 VvG stain |
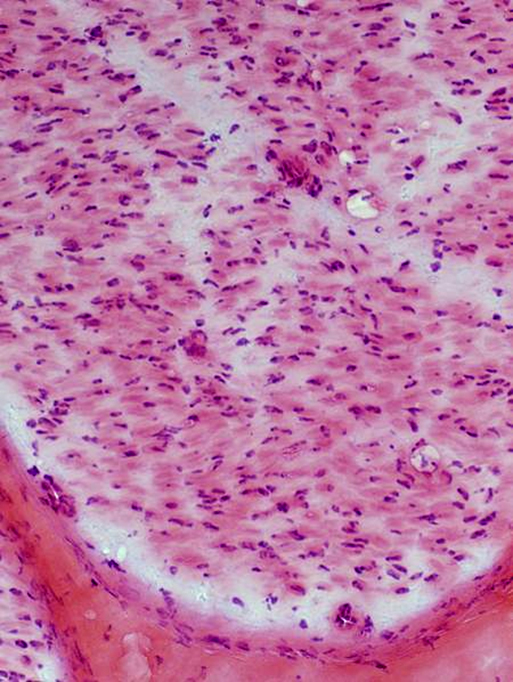
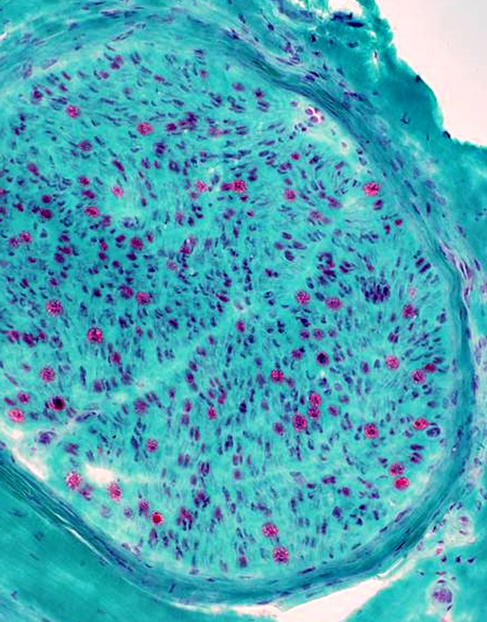 Gomori trichrome stain |
|
|
|
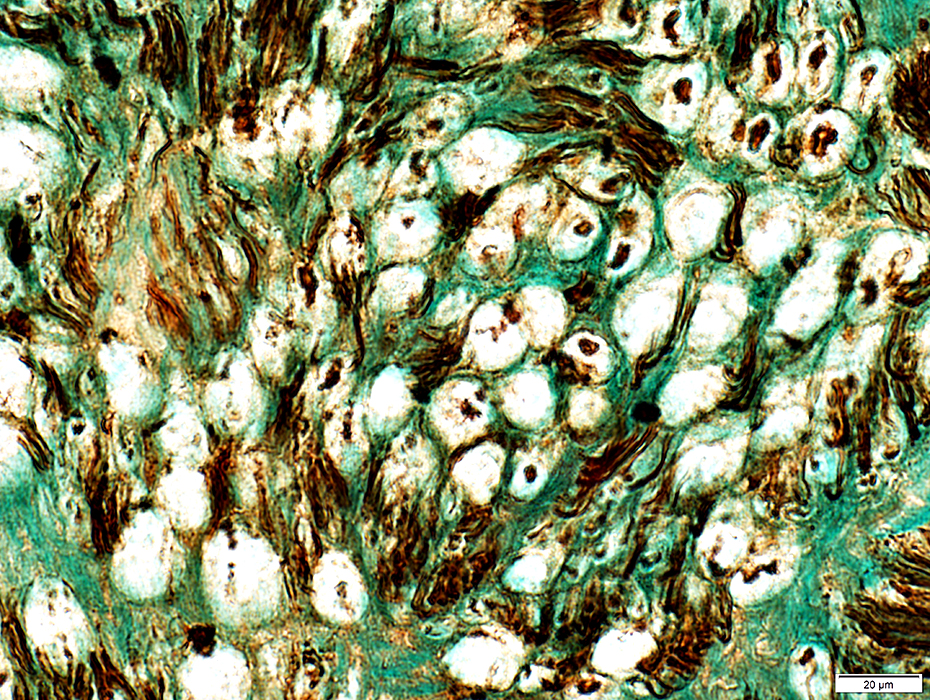
Myelinated Axon loss: Severe
| |
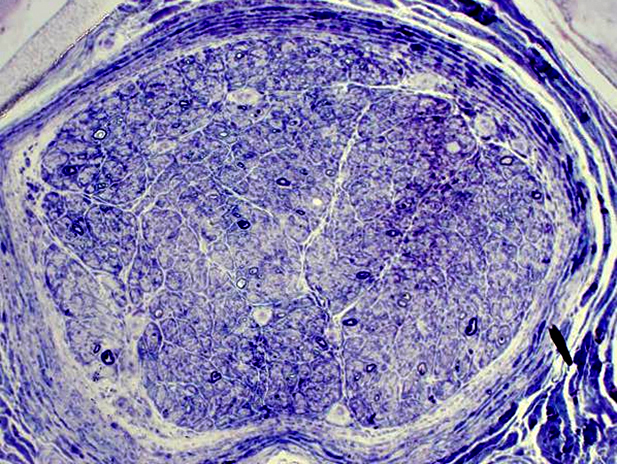
Axons, Large & Small: Comparative changes
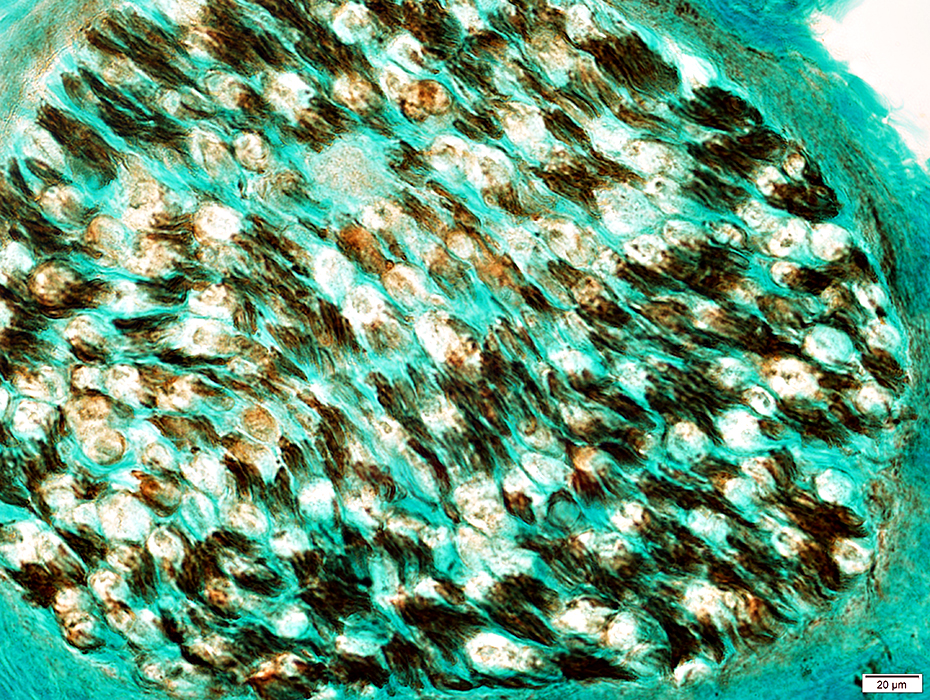
Myelinated Axon loss: Large > Small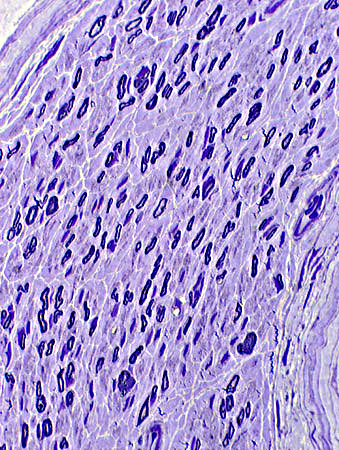 Toluidine blue stain
|
Myelinated Axon loss: Small > Large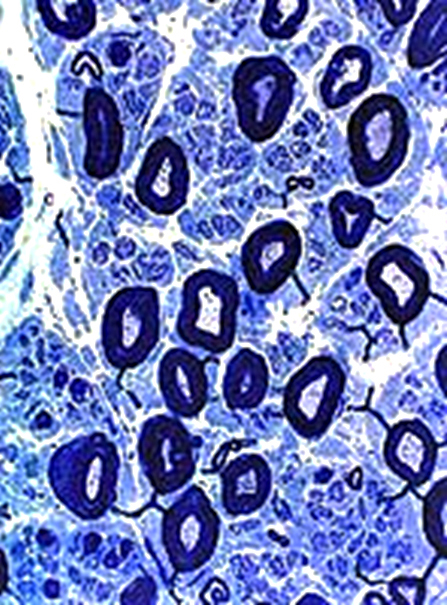 VvG stain |
|
Large axons: Moderately severe loss | |
|
Small Axons: Relatively preserved: Many Small axons, but Few large, myelinated axons Small axons are diffusely distributed, not clustered, in endoneurium 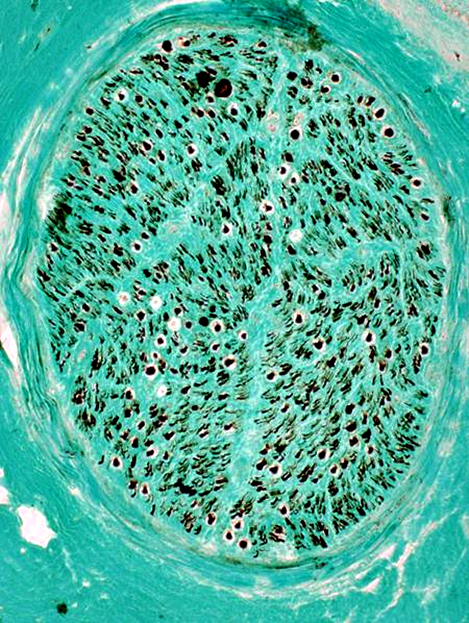 Neurofilament stain
|
Remaining Large Myelinated Axon (Arrow)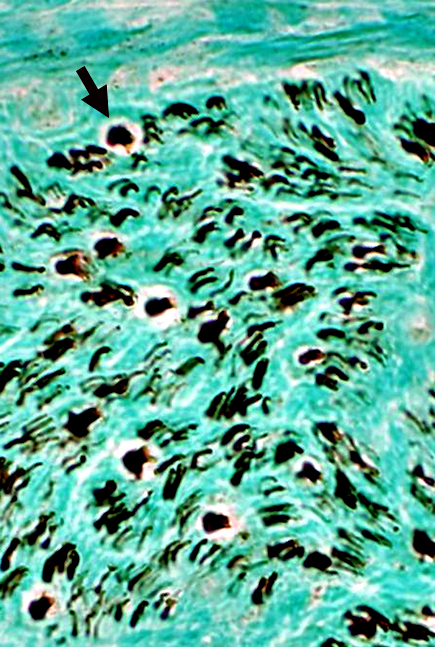 Neurofilament stain
|
Axon loss, severe: Large & Small axons are both markedly reduced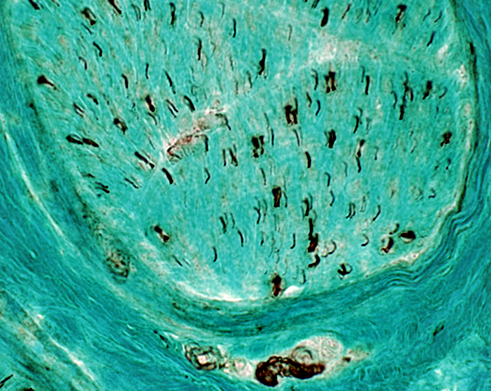 Neurofilament stain |
Myelinated axons: Severe loss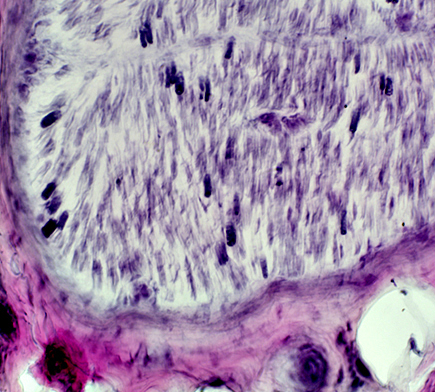 VvG stain |
Wallerian Degeneration 5
Wallerian Degeneration: Principles & Features
Alternate axon degeneration pathway: Trophic withdrawal induced
|
|
Wallerian Degeneration: Pathology
|
Axon loss Early Damage & Loss Relationship to myelin Ultrastructure Chronic Collagen pockets Bungner bands Regeneration Myelin Early Structure changes Degeneration Early Fragmented in Schwann cells Degradation Lipid & Myelin debris Ovoids Schwann cells Autophagic |
Waller illustration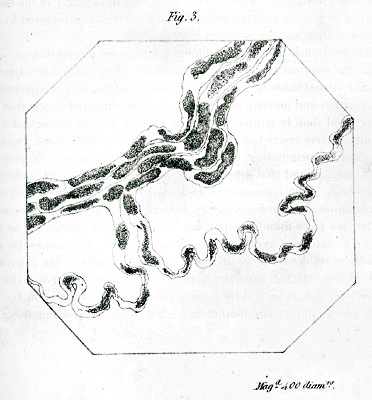  5 days after nerve section (Hypoglossal nerve) |
Axon Degeneration: Patterns & Morphology
Axons: Degeneration, Ongoing, Early
|
Axon Loss Histology Morphology/Ultrastructure Very Early Axoplasm loss Organelle aggregation |
Myelin Pathology Histology Periphery C5b-9 deposition MxA+ cells Myelin Ovoids/Remnants Schwann cells Autophagic Ultrastructure Inner layers Compact Outer layers Later stages Ovoids Autophagic Schwann cells Cells with Lipid debris |
Degeneration of Myelinated Axons: Ultrastructure
Axon Loss: Early
Neurofilament stain of Axons: Reduced or AbsentNeurofilament-stained axons: Lost within regions of myelin (MBP stain)
Axons: Neurofilament stain
Reduced or Absent staining for Large axons (Yellow)
Staining for Small axons (Green) remains
Mechanism of axon loss: SARM1
Myelin-Basic Protein (MBP) (Red): Present in remaining myelin
Regions with MBP stain are abnormal: Costain with NCAM
Also see: Control Nerve
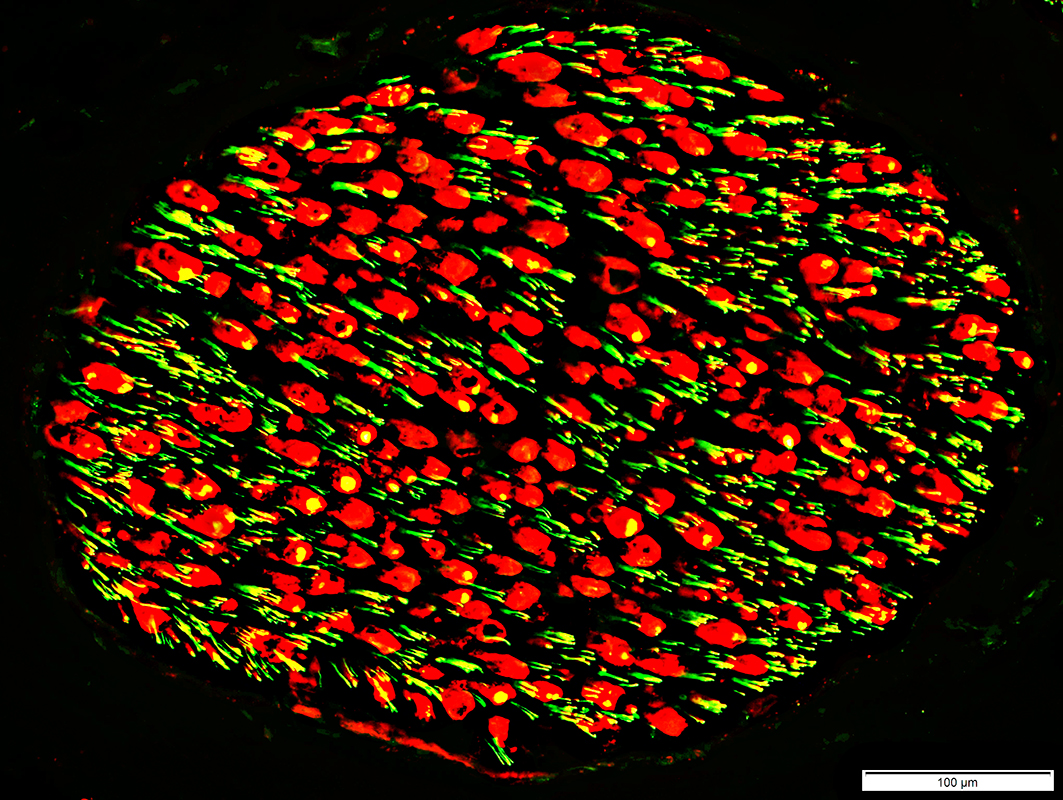 Neurofilament stain (Green or Yellow); Myelin Basic Protein stain (Red) |
Control Nerve
Myelin basic protein (Red) surrounds large axons (Yellow)
Normal clusters of small unmyelinated axons (Green)
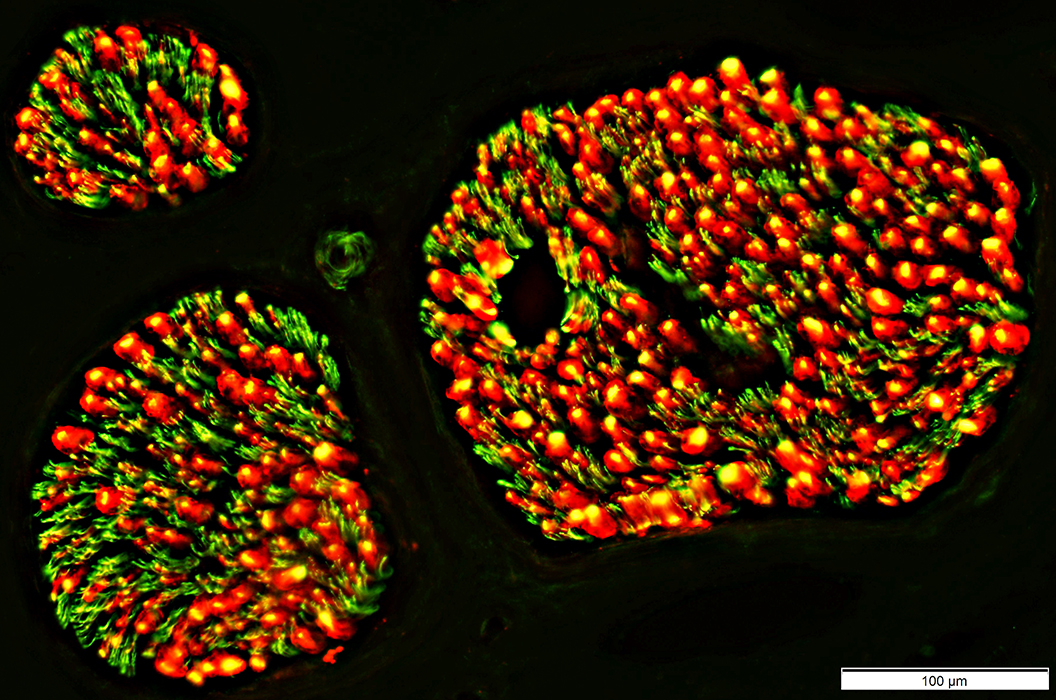 Neurofilament stain = Green; Myelin basic protein stain = Red |
Control nerve
Moderately later pathology
Large Axon loss: Chronic
 Neurofilament stain |
|
Loss of Large Axons: Active Neurofilament staining: Areas that normally contain axons & myelin have Absent staining of large axons Neurofilament debris: Granules stained NCAM staining Myelin debris/ovoids: Abnormally stained for NCAM Small, Unmyelinated, Axons Neurofilament staining: Normal clusters Compare to: Control nerve |
 Neurofilament stain |
 NCAM stain |
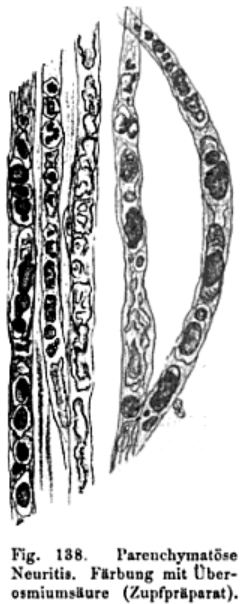
Myelinated Axon Degeneration: Very Early
Axon Pathology
 From: R Schmidt |
WD Very Early: Axoplasm Aggregates
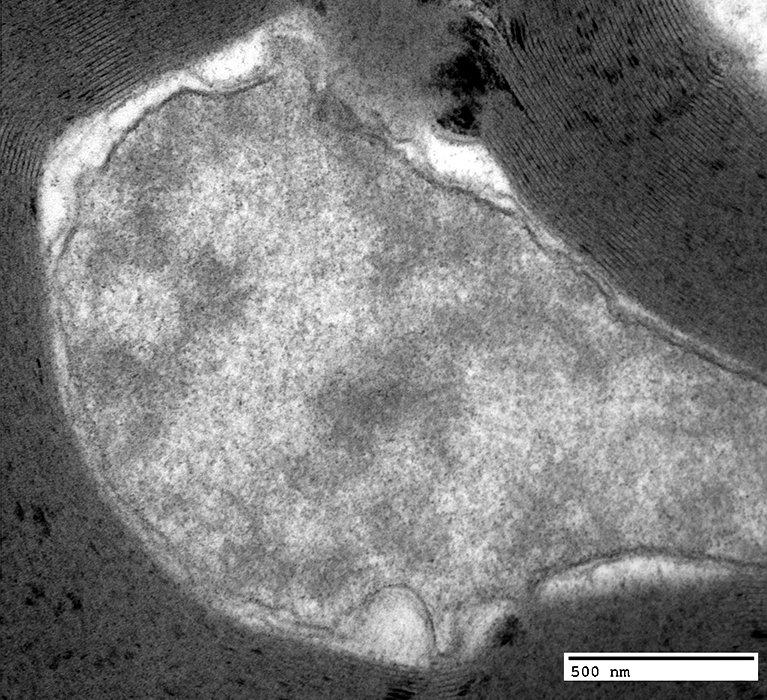
|
WD Very Early: Axoplasm: Dark & Homogeneous
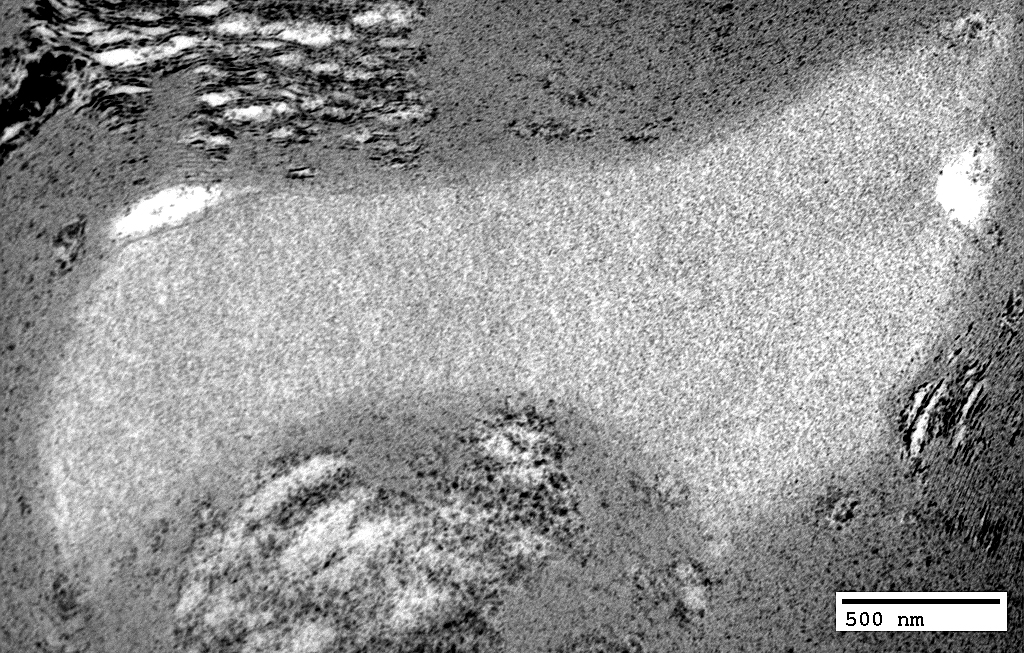
|
WD Very Early: Axoplasm
Pale
Few organelles
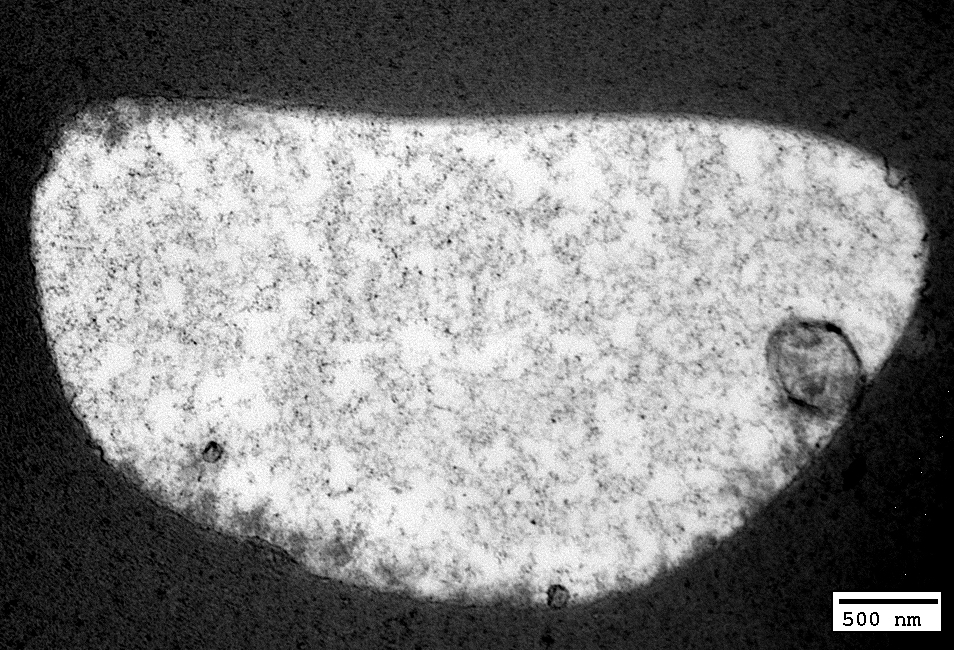
|
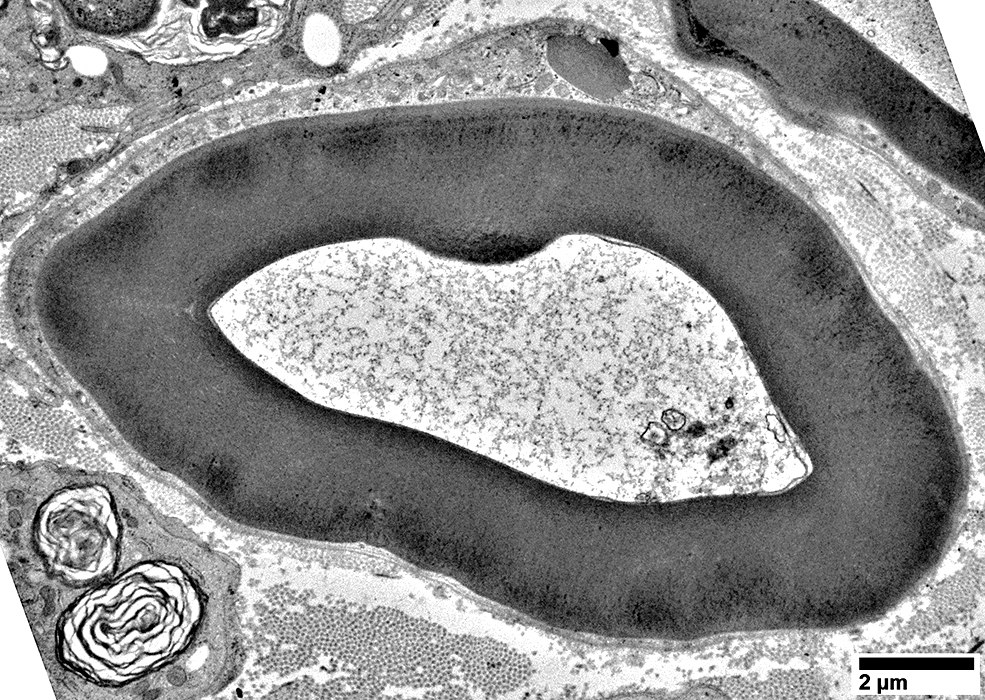
|
Aggregated organelles
Clustered regionally in axon: Near mildly abnormal myelin structure
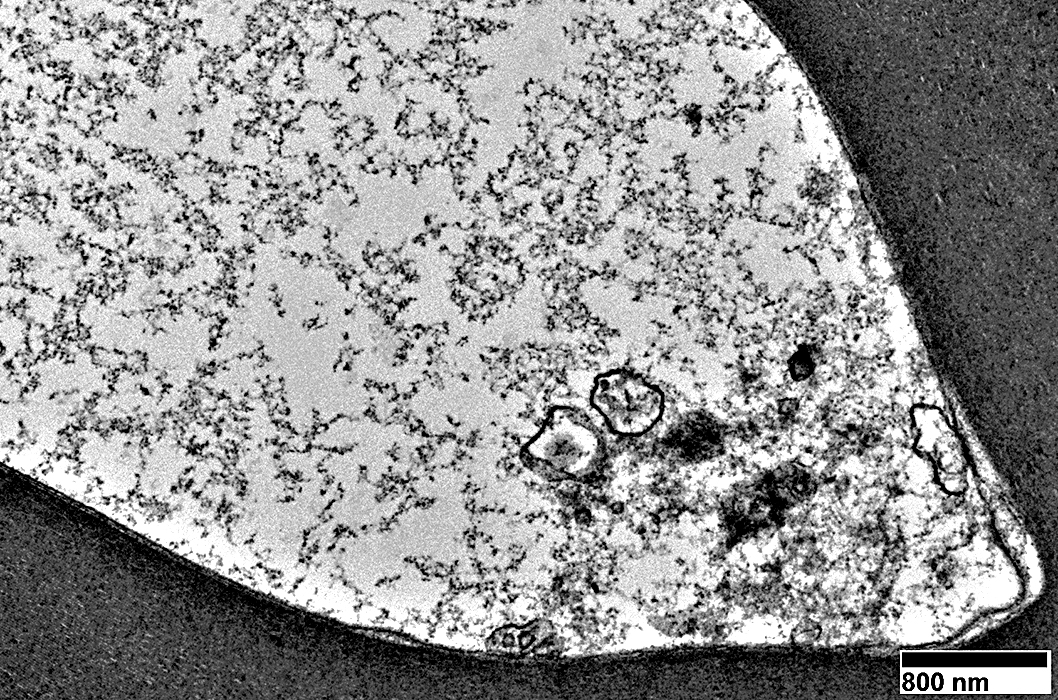
|
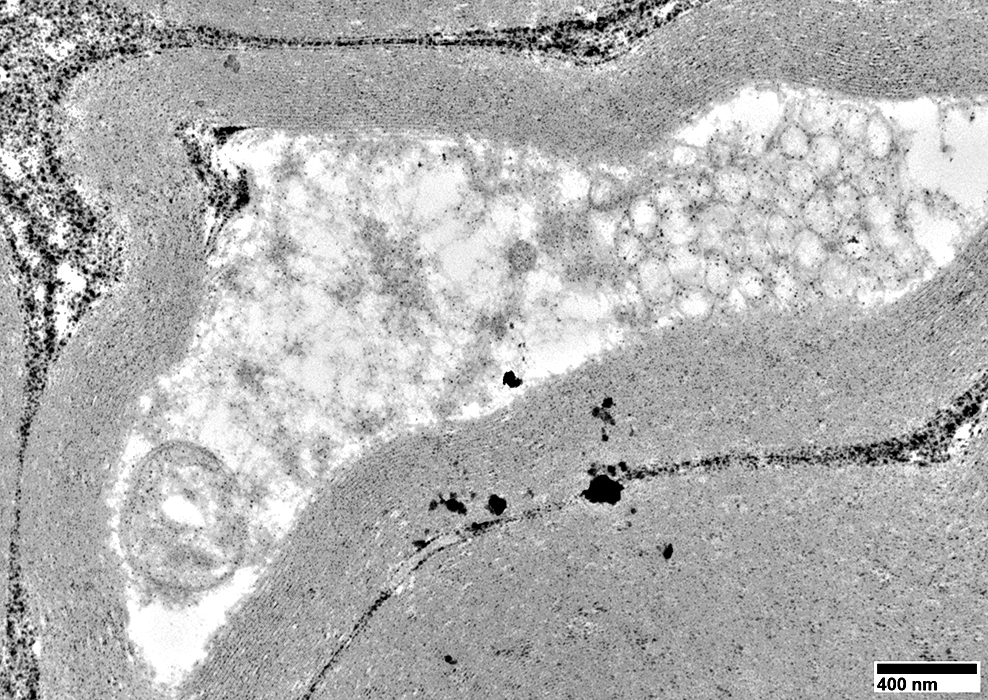 From: R Schmidt |
Volume: Reduced
Structure
Irregular or Pale
May contain vesicles or organelles
Myelin sheath: May appear very thick compared to axon size
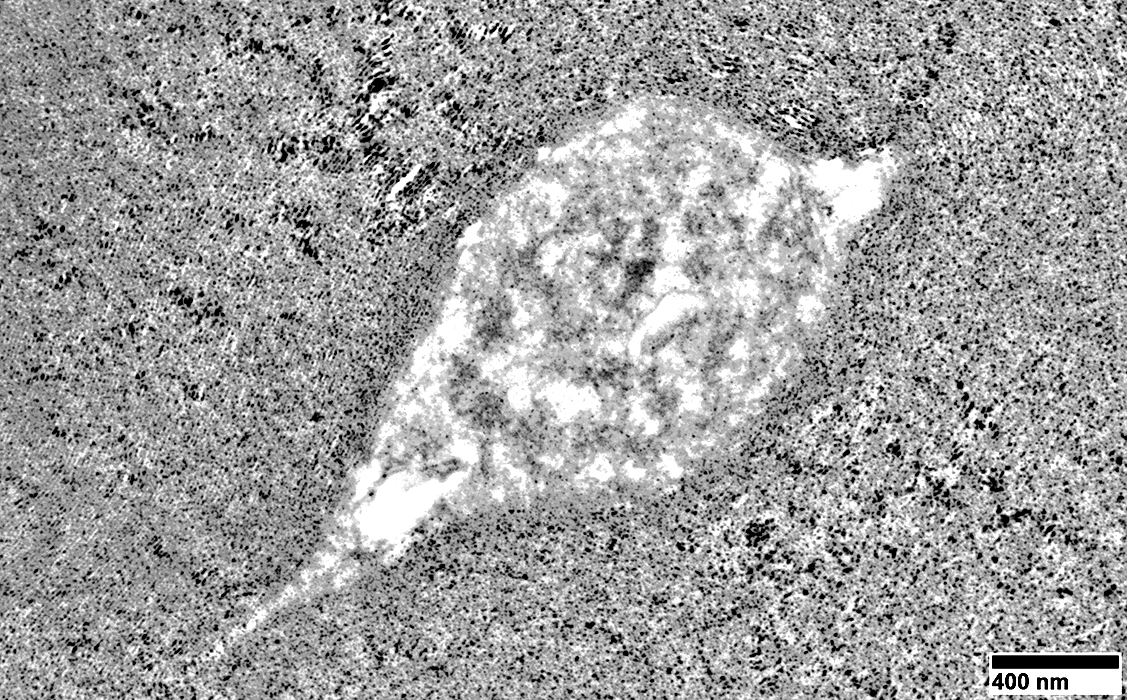 From: R Schmidt |
 From: R Schmidt |
Volume: Reduced
Structure
Irregular or Pale
May contain vesicles or organelles
WD Moderately Early: Axoplasm loss & Pathology
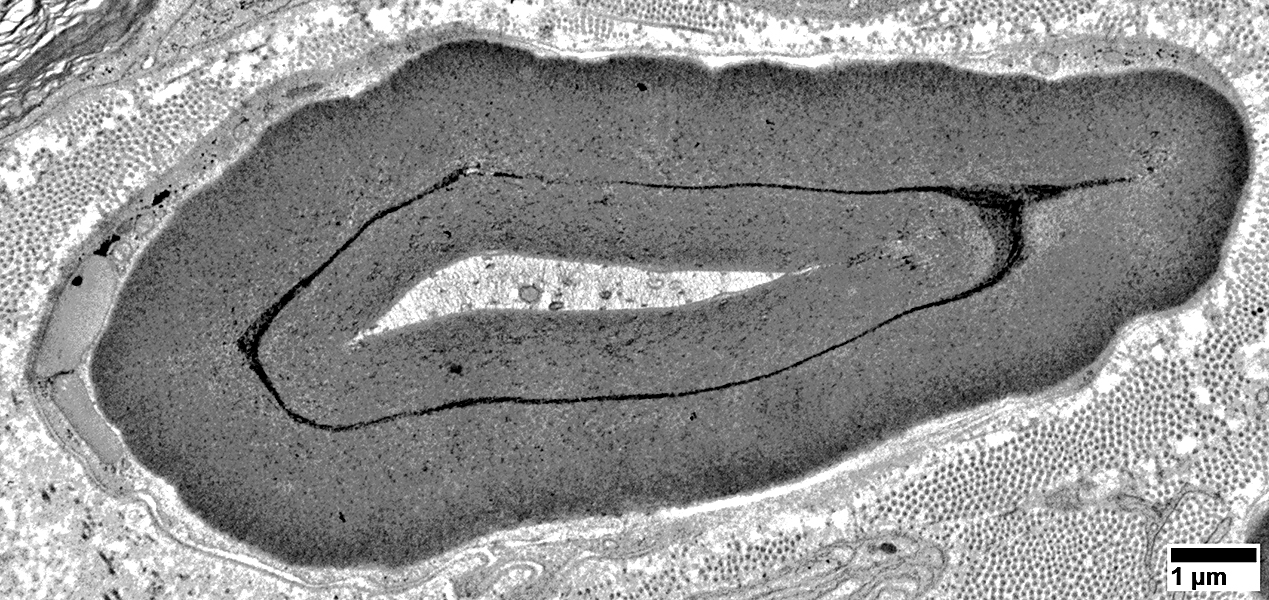 From: R Schmidt |
Volume: Reduced
Structure: May be irregular
Myelin sheath: Appears very thick compared to axon size
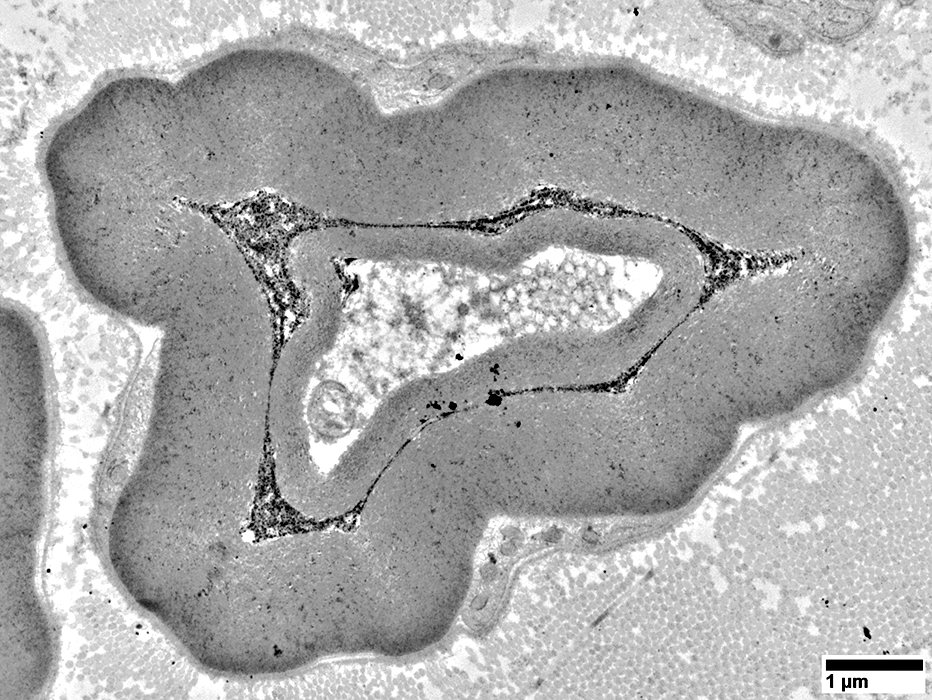 From: R Schmidt |
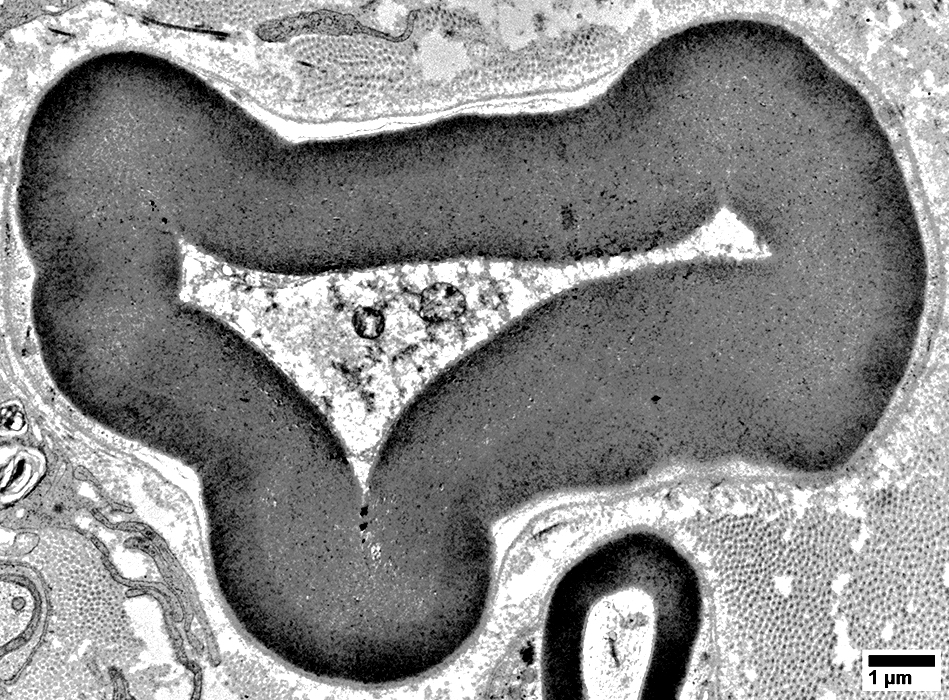 From: R Schmidt |
Volume: Reduced
Structure: May be irregular
Myelin sheath: Appears thick compared to axon size
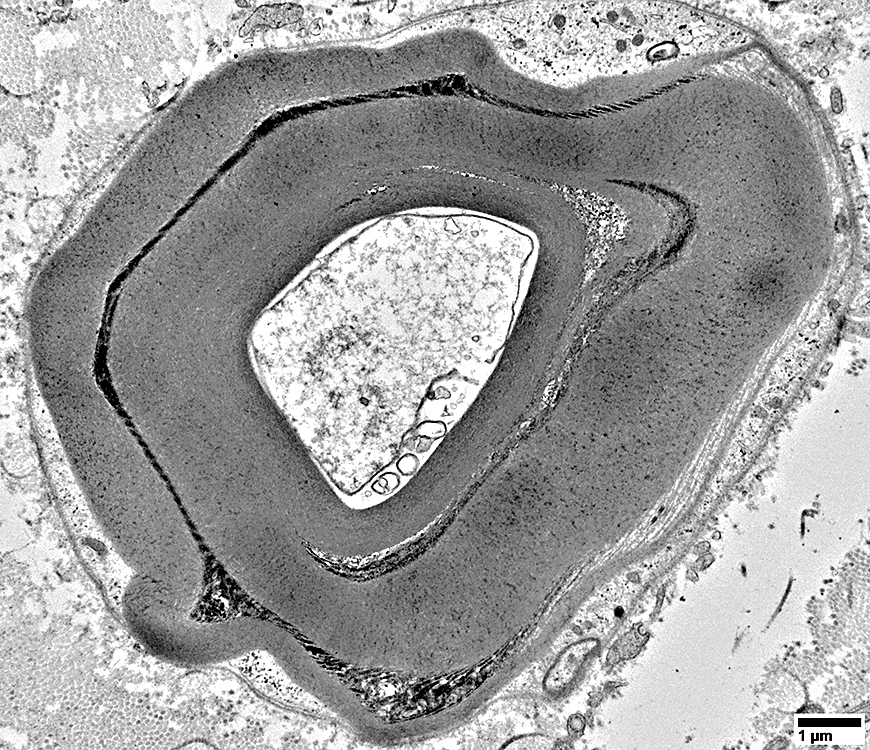 From: R Schmidt |
Axoplasm
Blebs: May be present between damaged axon & myelin (Below)
Myelin ± Axon Pathology: Early
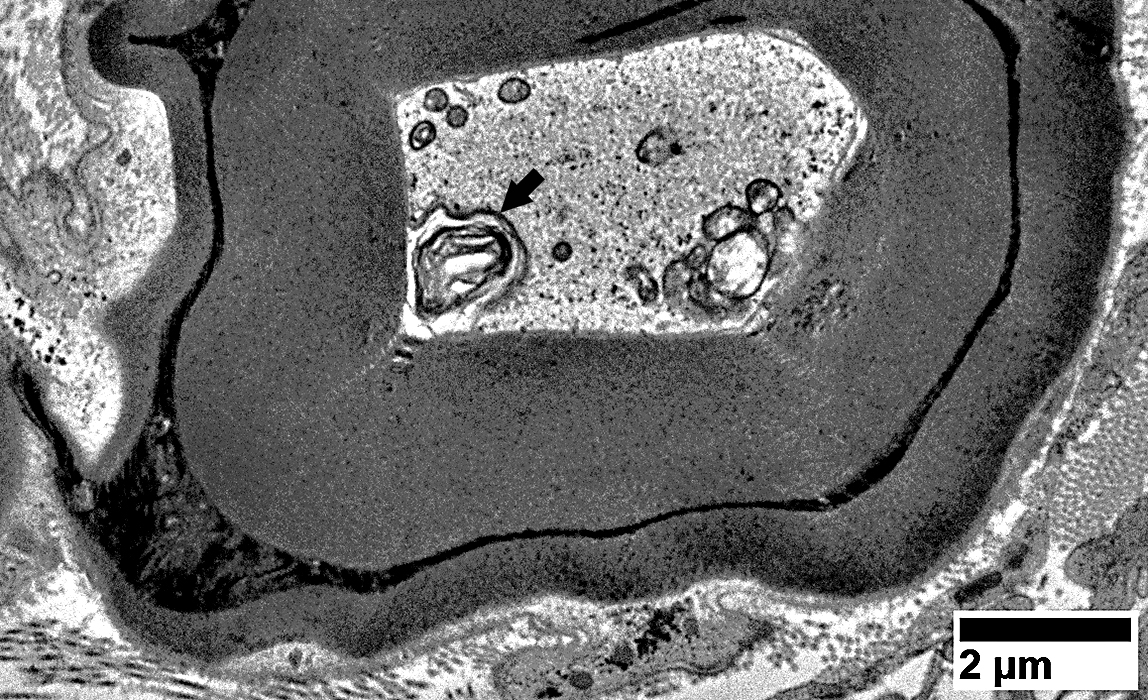 From: R Schmidt |
Clustered organelles
Myelin: Early WD Changes
Internal Pathology: Bleb, from internal layers of myelin sheath, indents axon
Sheath: Thick compared to axon size
WD Very Early
Myelin
Abnormal structure of internal layers
Axon
Aggregated organelles
Clustered regionally in axon: Near abnormal myelin structure
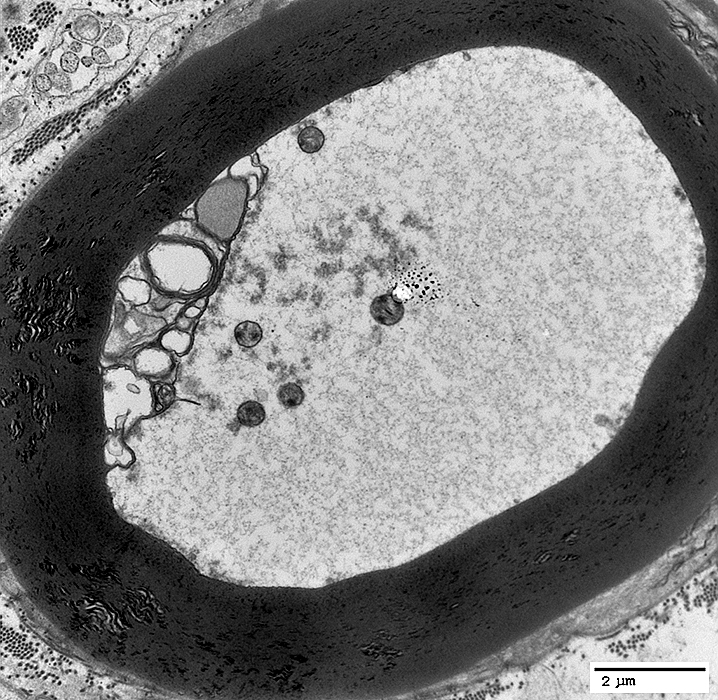
|
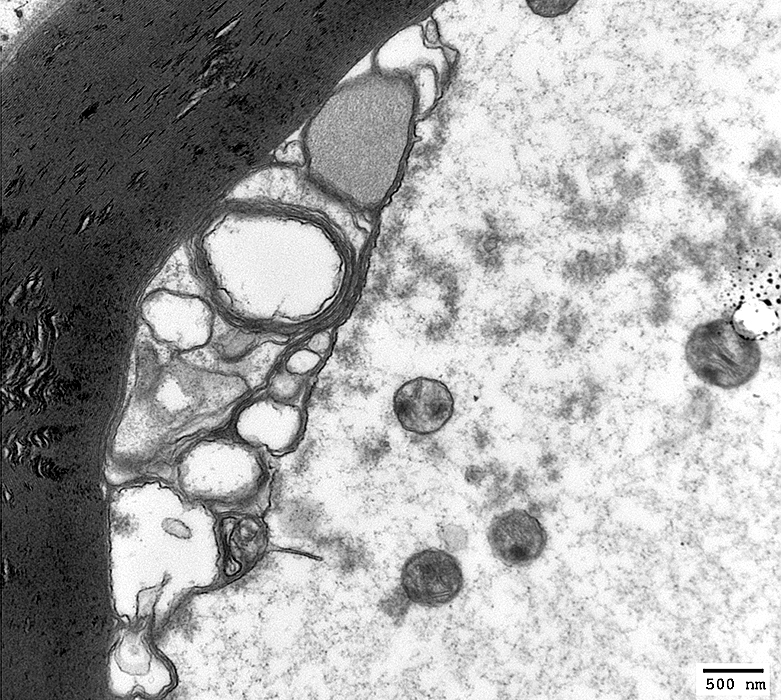
|
Axon
Aggregated Organelles
Remnants of axon within remaining compact myelin
Myelin: Irregular internal areas
No associated phagocytic cells
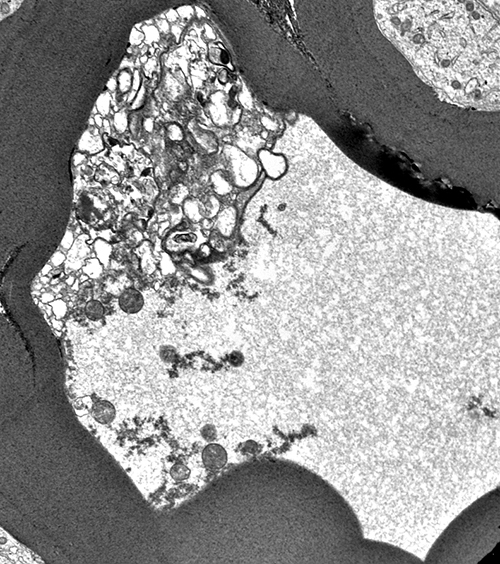 From: Robert Schmidt MD |
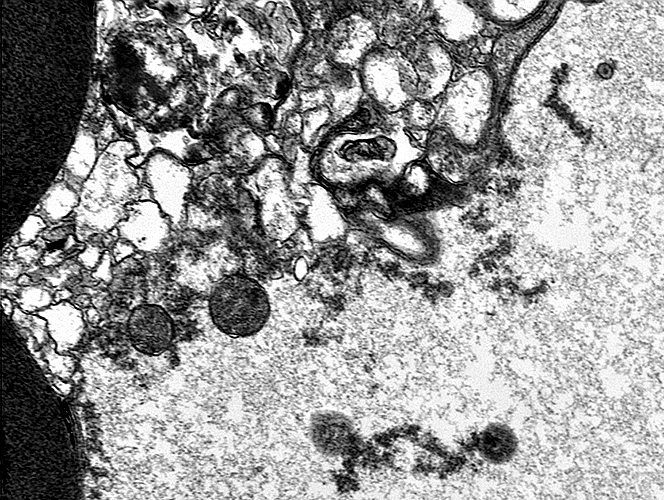 From: Robert Schmidt MD |
WD Early
Myelin: Blebs from inner myelin layers indent axons
|
Compact Myelin: Abnormal Structure
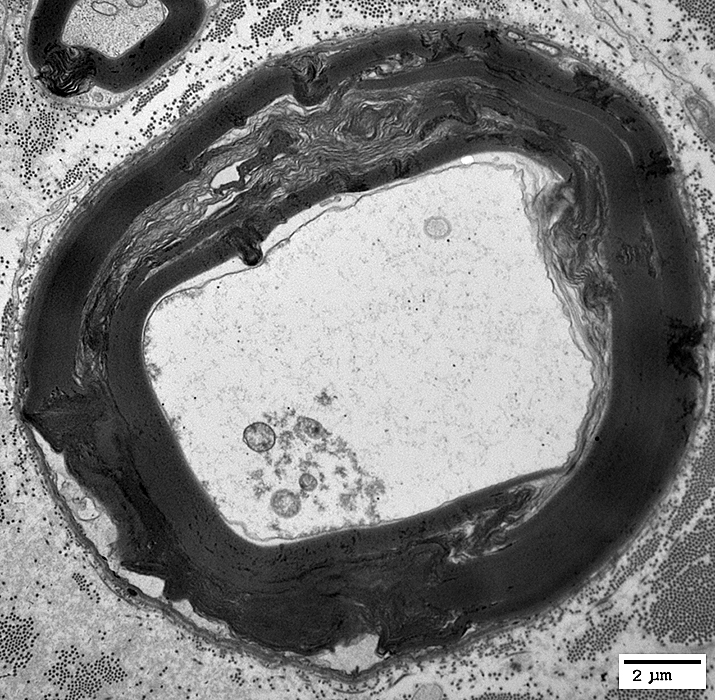
|
Axoplasm is pale
Organelle aggregates are present
Myelin Early WD Changes
Structure of compact myelin is disrupted
Outer Myelin Layers: Abnormal Structure
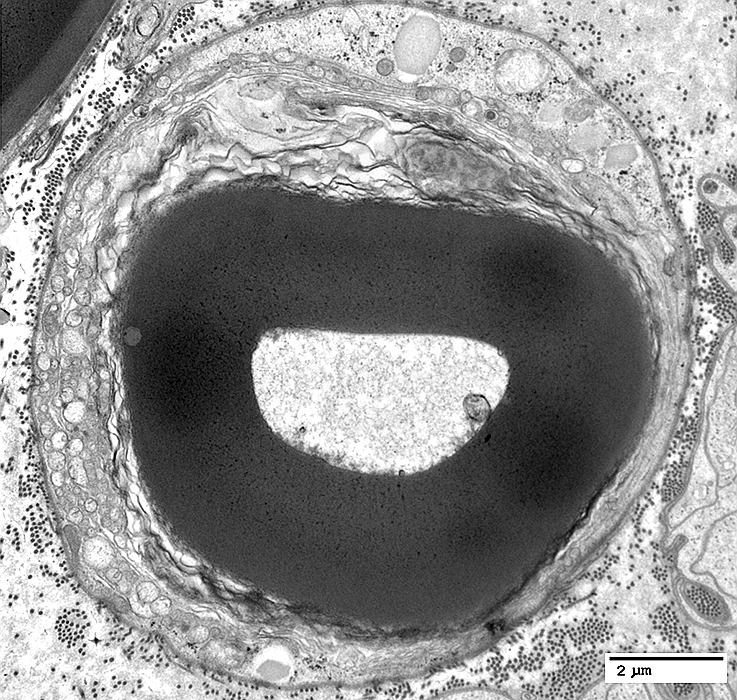
|
May be: Pale or Aggregated
Myelin Early WD Changes
Myelin outer layers: Abnormal structure
Remaining myelin: Thick compared to axon size
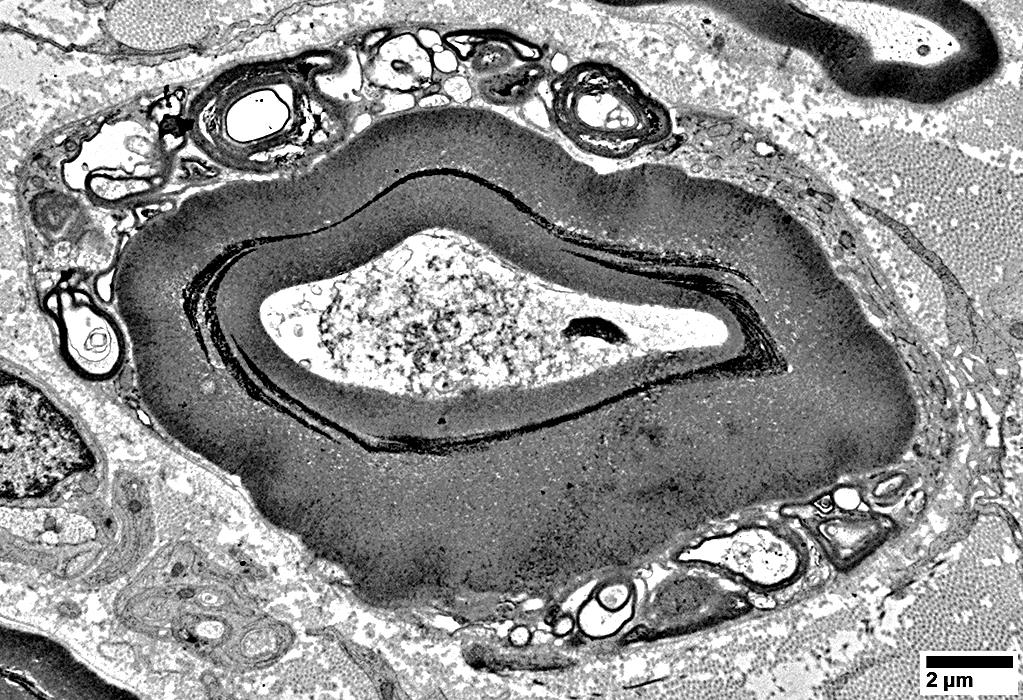 |
WD, Early: Ab-Axonal Schwann cell Cytoplasm
Pale background
Irregular aggregates
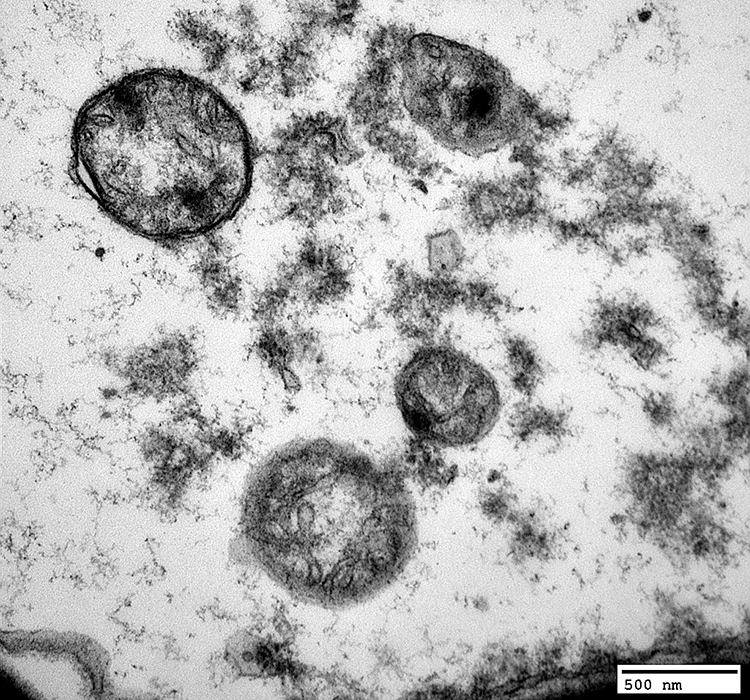
|
Wallerian Degeneration
Wallerian Degeneration, Days to Few WeeksRegions of Myelin basic protein (Red) often have no associated axons
Loss of small unmyelinated axons (Green)
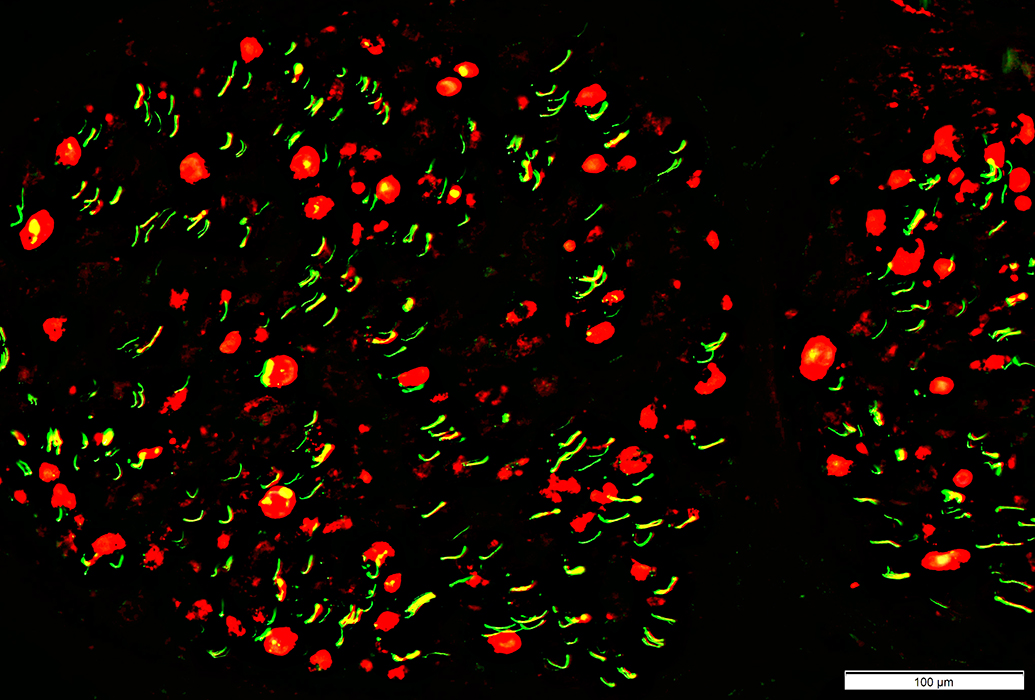 Neurofilament stain = Green; Myelin basic protein stain = Red |
Loss of Large Axons: Moderately later than above
Axons (Yellow & Green; Neurofilament stain)
Absent, or Reduced large axons: Axons inside Myelin basic protein stained myelin (Schwann cells)
Areas where axons are lost are black
Small axons are relatively preserved
Myelin basic protein stain (Red)
Abundant
Shapes: Variable and irregular
No associated axons
Central dark areas where axons are lost
Regions of Myelin Basic Protein (Red) have no associated Neurofilament-stained (Yellow) axons
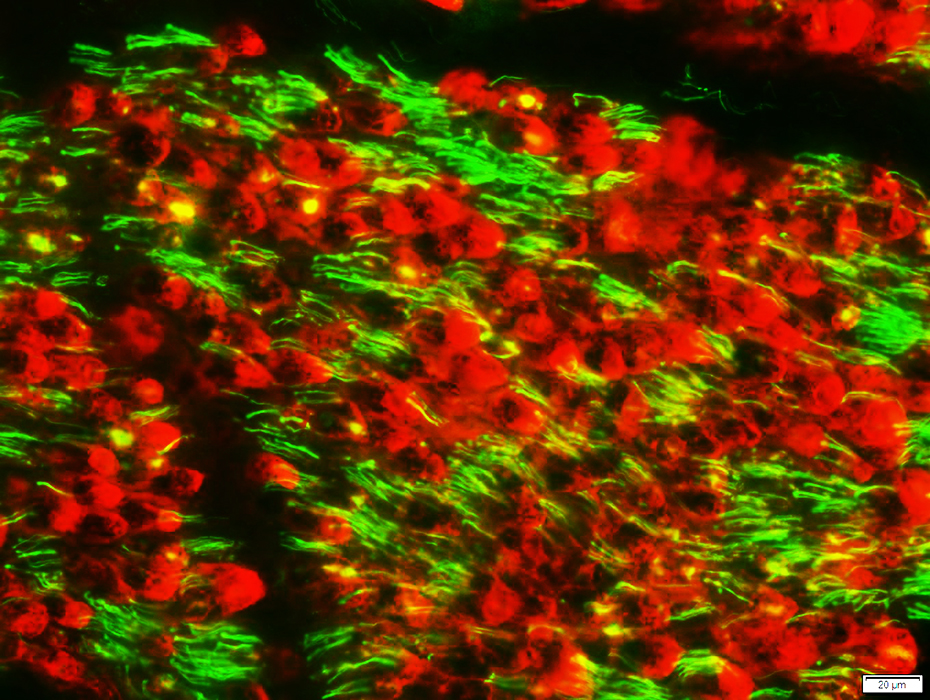 Neurofilament (Green) & Myelin basic protein (Red) stain |
Control nerve for comparison
Axon loss: Earlier
Axon Loss: Myelin Pathology
 H&E stain |
Stain: Reduced or lost
Structure: Lost or clumped
 Gomori trichrome stain |
 VvG stain |
 H&E stain |
Stain: Reduced or lost
Structure: Lost or clumped
 Gomori trichrome stain |
 Gomori trichrome stain |
Myelin is still present
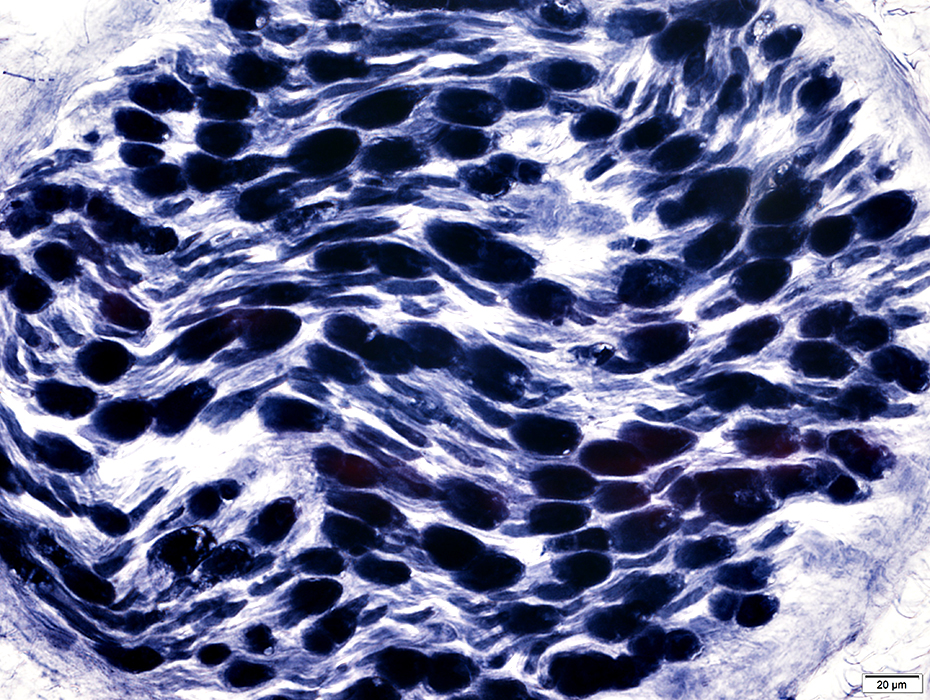 VvG stain |
Many Myelin sheaths are paler stained
 VvG stain |
Wallerian Degeneration: Schwann cells, Myelinating: Molecular pathology
NCAM expressed in myelin sheath
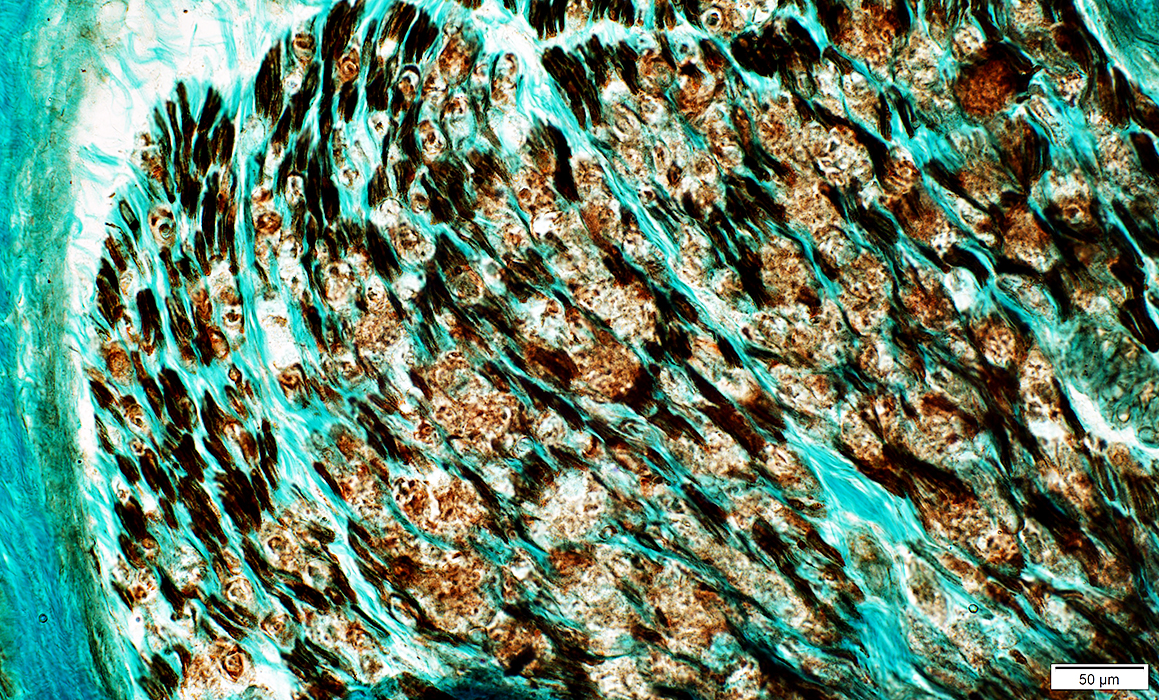 NCAM stain |
Normal (Below): NCAM expressed mainly in adaxonal cytoplasm
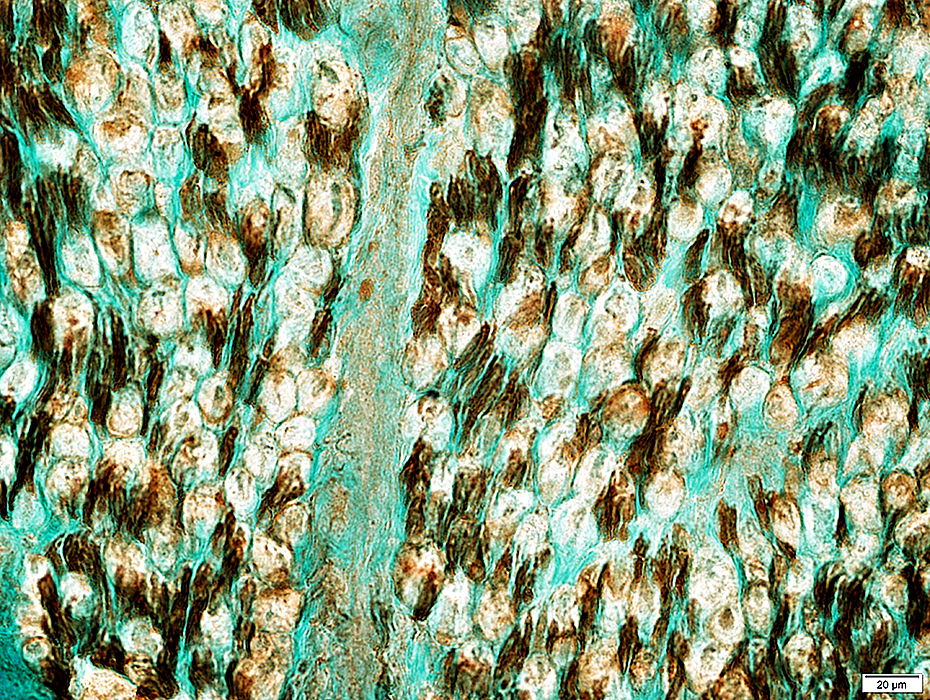 NCAM stain |
Normal (NCAM+) Non-myelinating Schwann cells
Present in clusters between myelinated axons
Not present in most regions of myelin sheath
Also see: Large axon loss, Chronic
Abnormal Co-localization of NCAM & MBP in myelin remnants (Yellow)
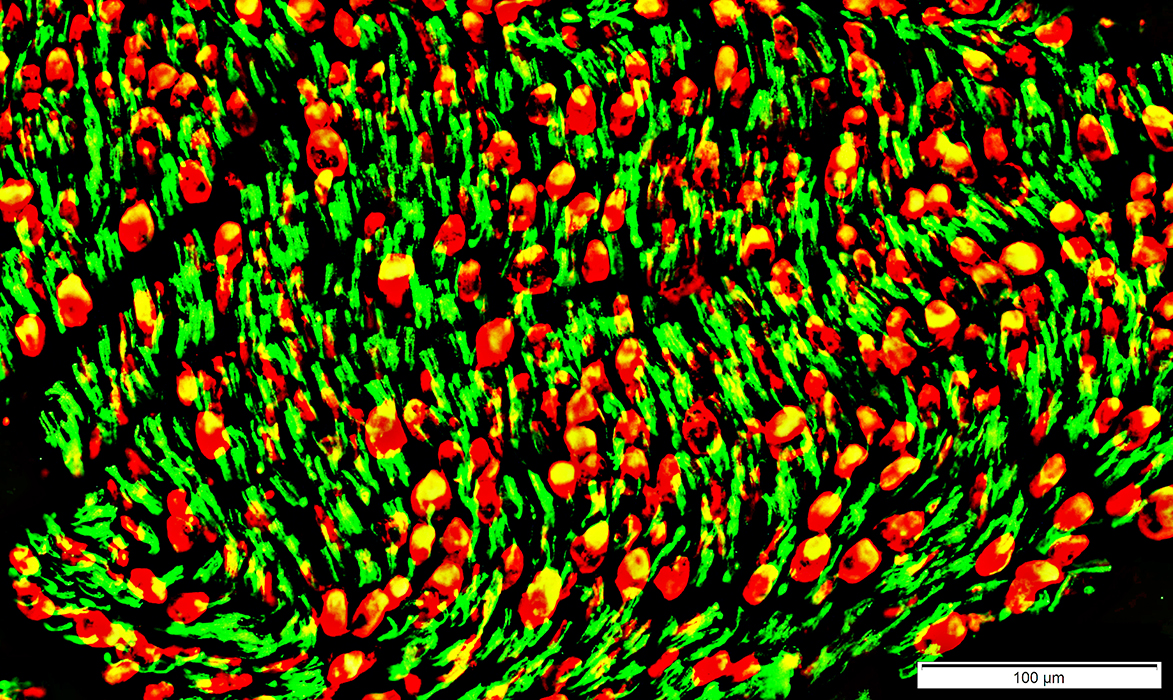 NCAM (Green) + Myelin basic Protein (MBP) (Red) stain |
Autophagic Schwann cells
 Acid phosphatase stain |
Fragmented myelin
Scattered endoneurial histiocytes
 Acid phosphatase stain |
Myelin & Axon degeneration: Ongoing, Early
Fixed nerve
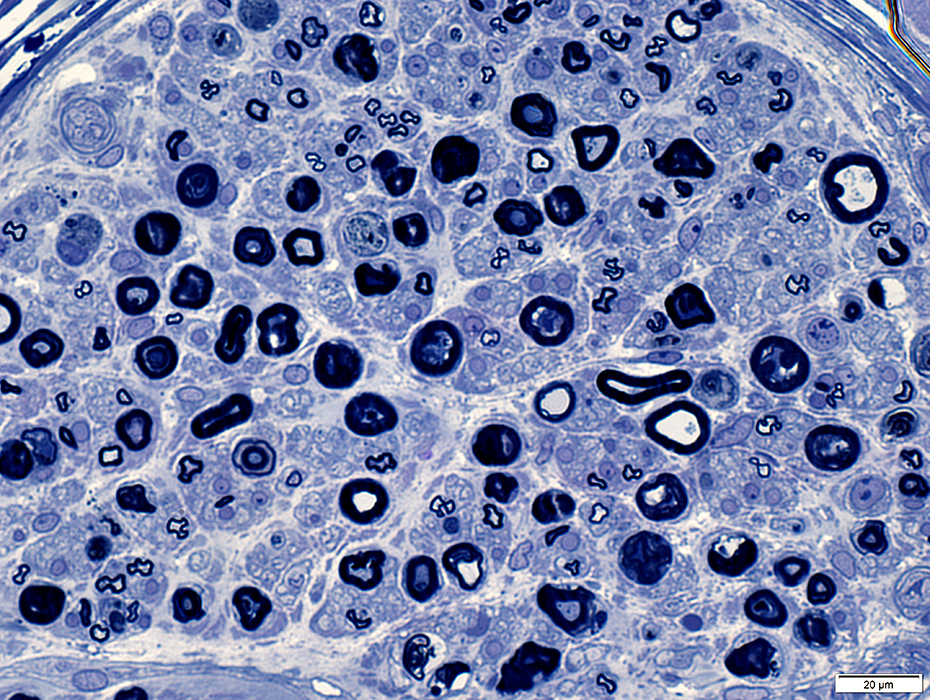 Toluidine blue stain |
Irregular, pale or dark, central regions
No phagocytes or fragmentation
Irregular morphology
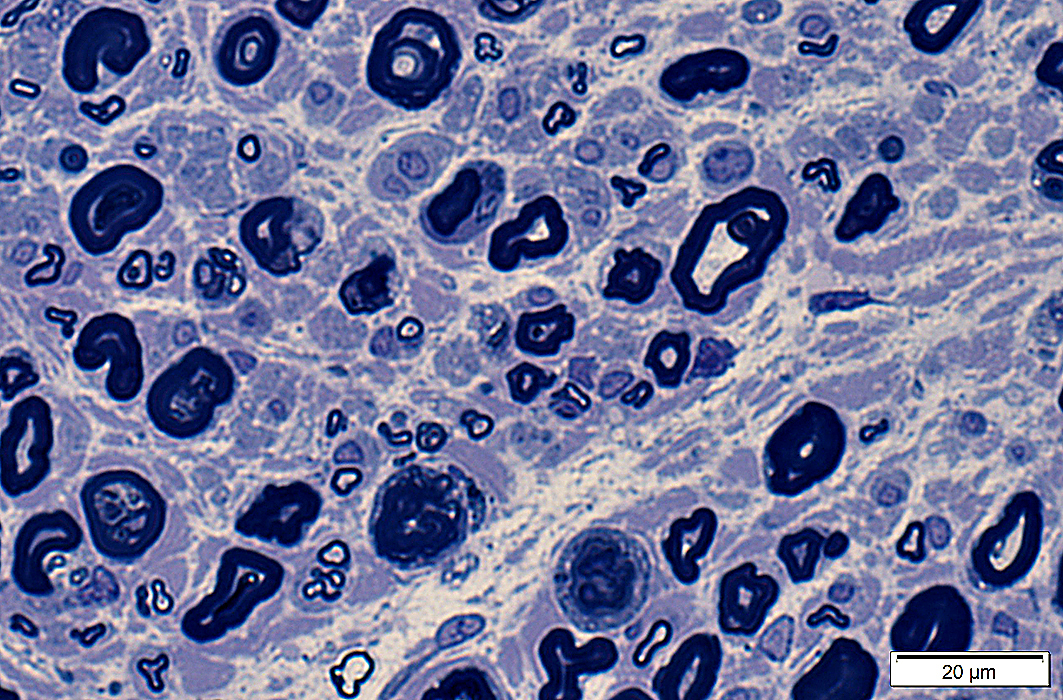 Toluidine blue stain |
Irregular myelin figures
Some remaining axons have dark axoplasm
No histiocytes (with lipid droplets in cytoplasm)
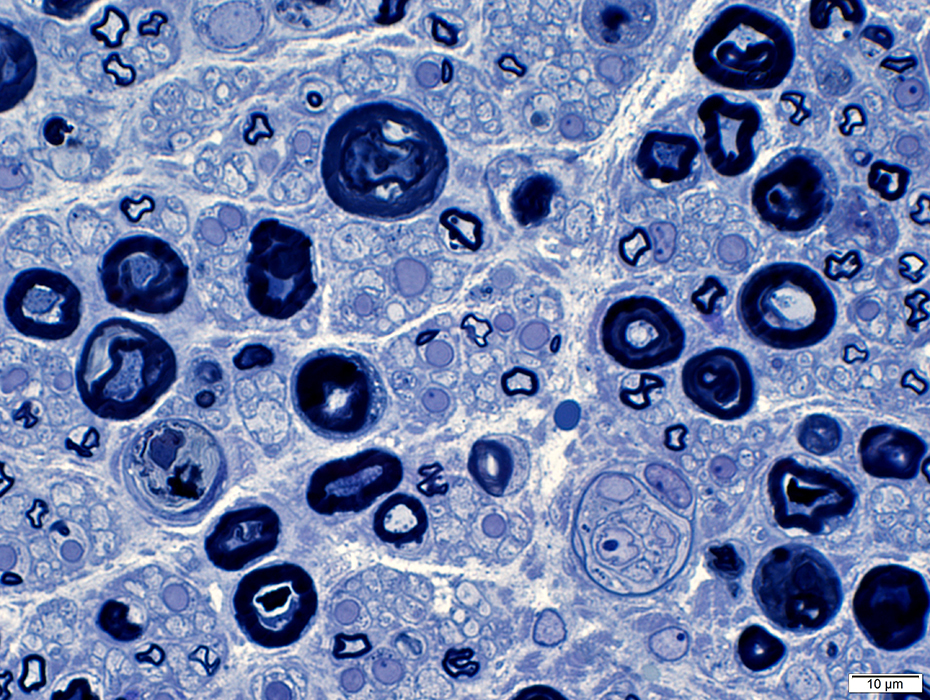 Toluidine blue stain |
Wallerian degeneration: Schwann cell pathology
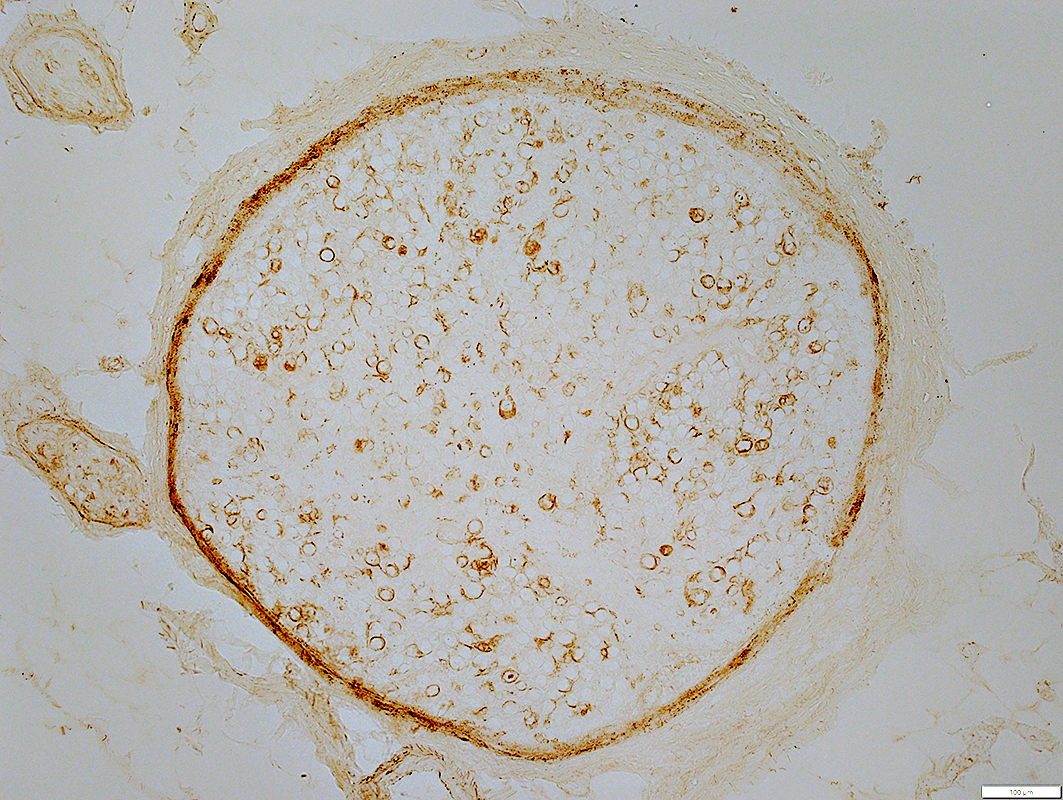 C5b-9 stain |
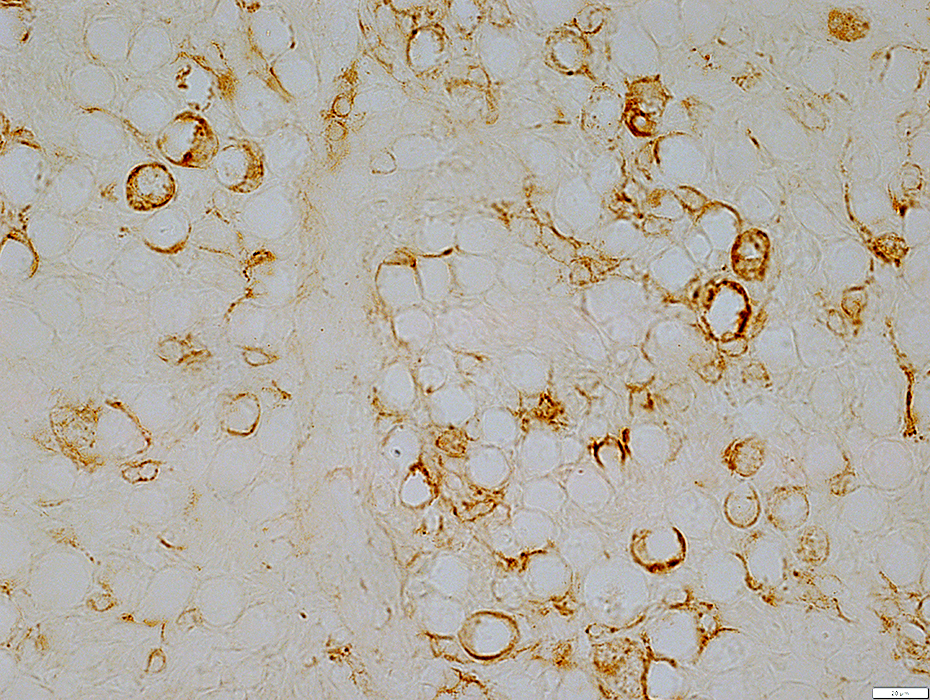 C5b-9 stain |
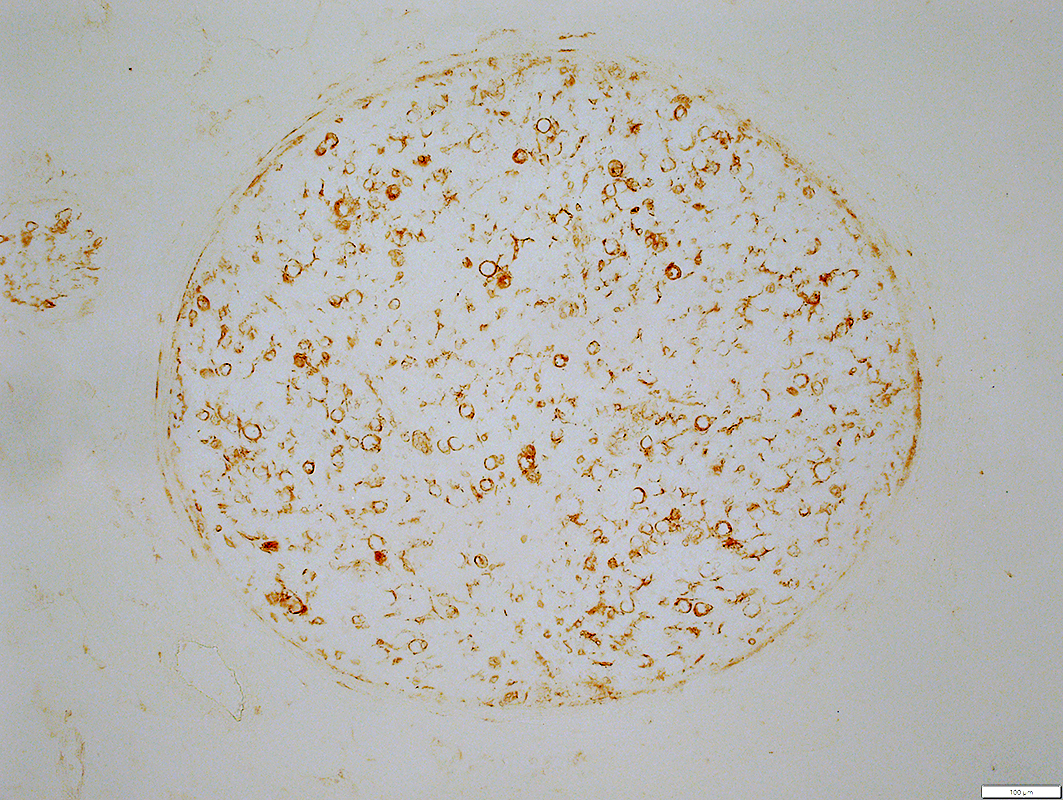 MxA stain |
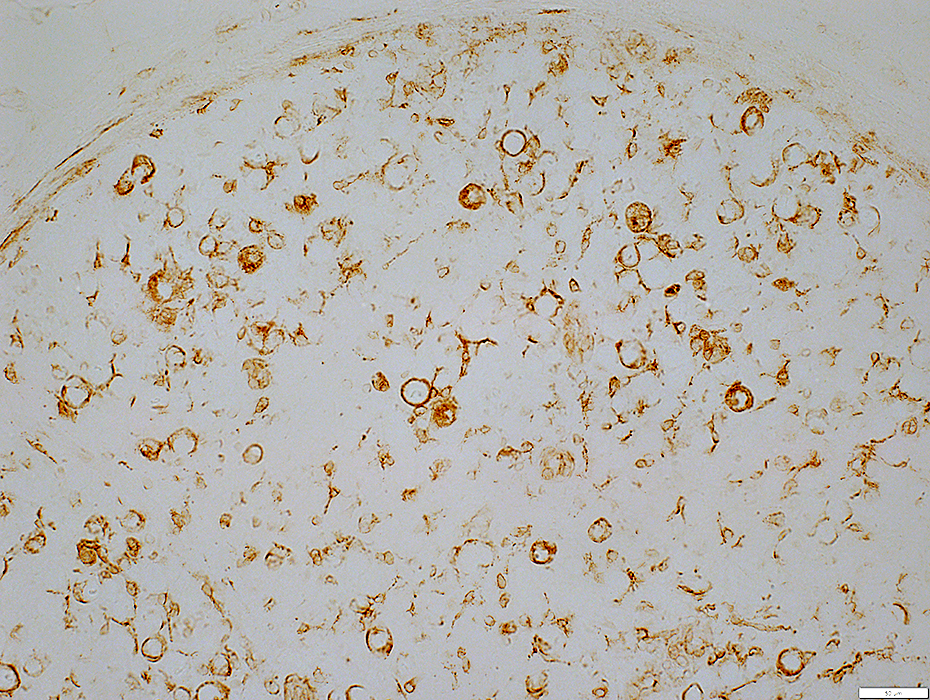 MxA stain |
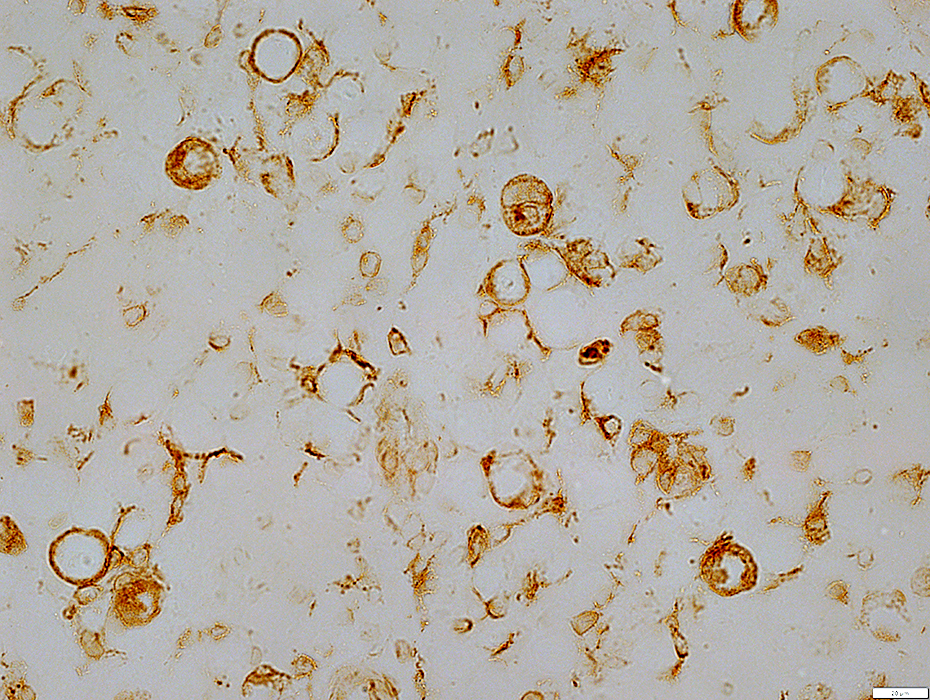 MxA stain |
Wallerian Degeneration: Intermediate stages
Myelin Breakdown: Fragmentation & Degeneration in Schwann cells
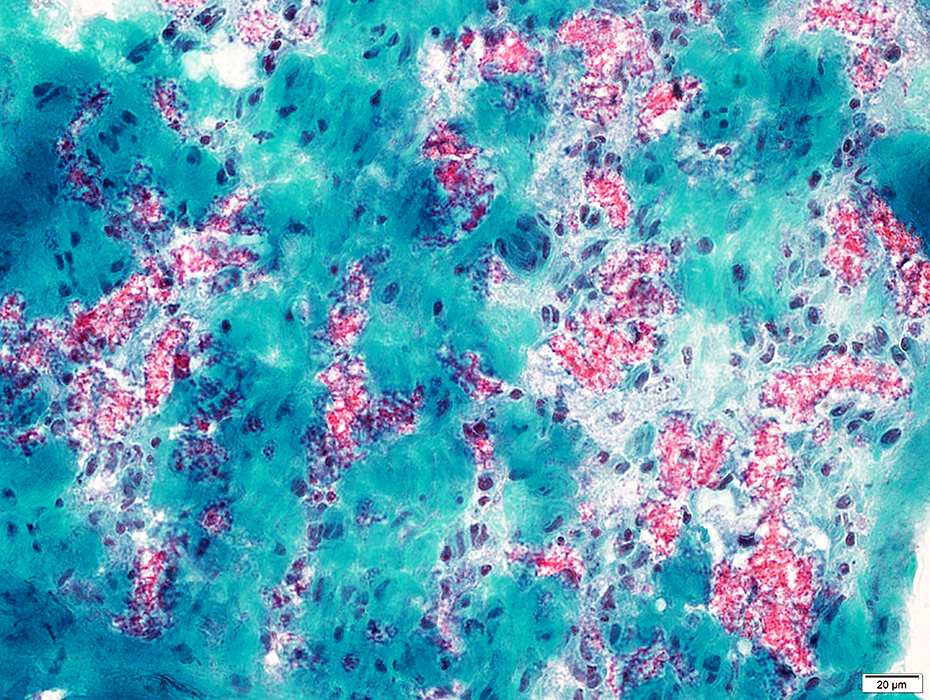 Gomori trichrome stain Wallerian degeneration: Early; Myelin fragmentation Myelin in phagocytic, post-myelinating cells: Clustered red (GT) or Black (VvG) endoneurial stain Also see: Normal control nerve |
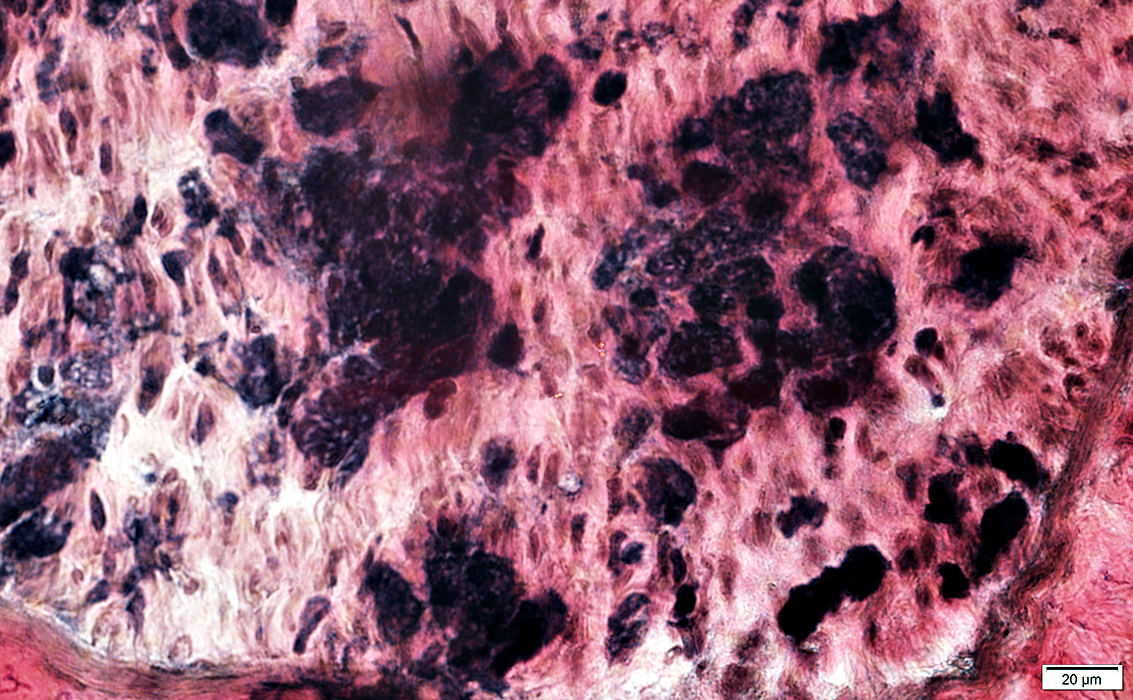 VvG stain |
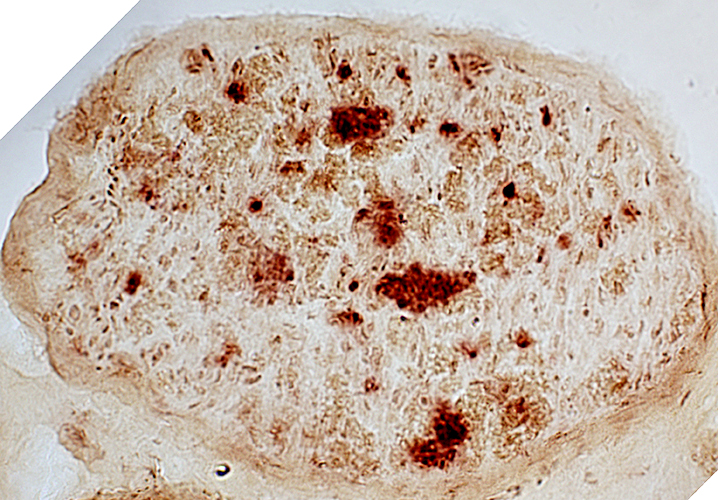 Acid phosphatase stain |
Large endoneurial cells (red) contain prominent lysosomal activity
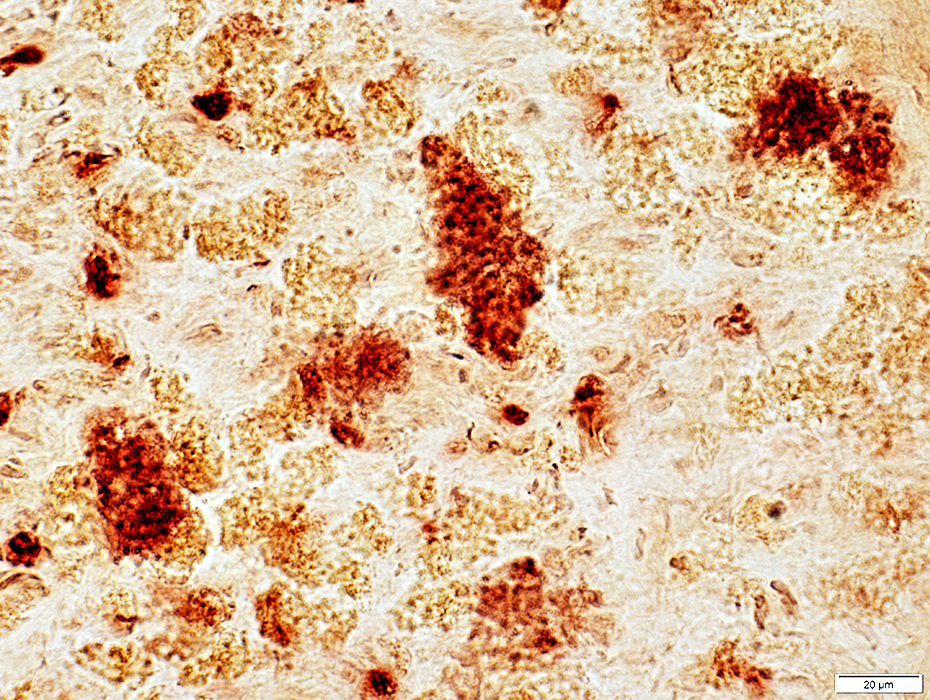 Acid phosphatase stain |
Large, Post-myelinating (Autophagic) Schwann cells
Autophagic
Contain
Fragmented myelin (Light arrow): In various shapes & stages of degeneration
Lipid droplets (Dark arrow): Small, Round & Clear
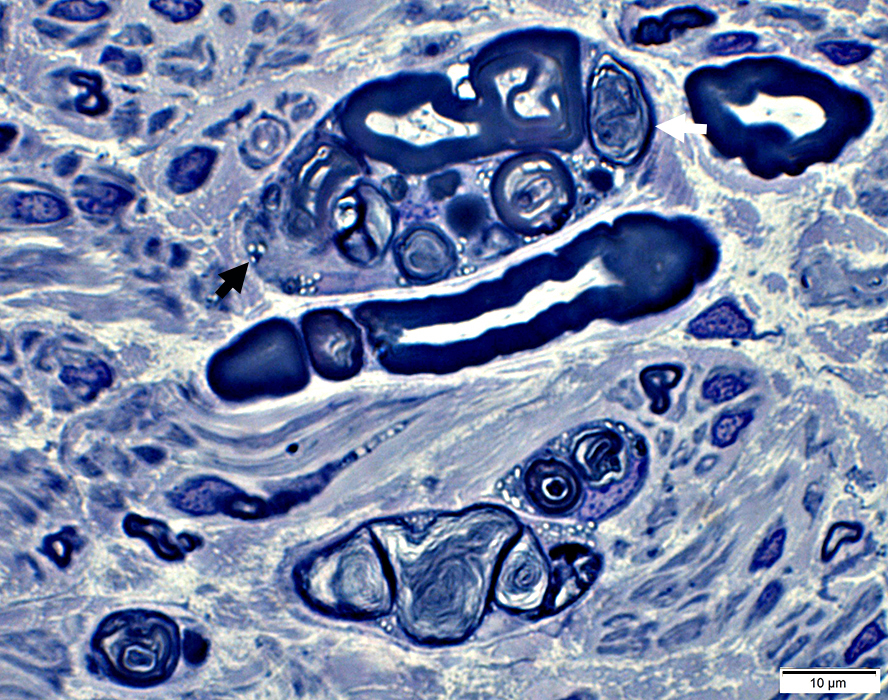 Toluidine blue stain |
Myelin fragmentation in Post-myelinating (Autophagic) Schwann cells
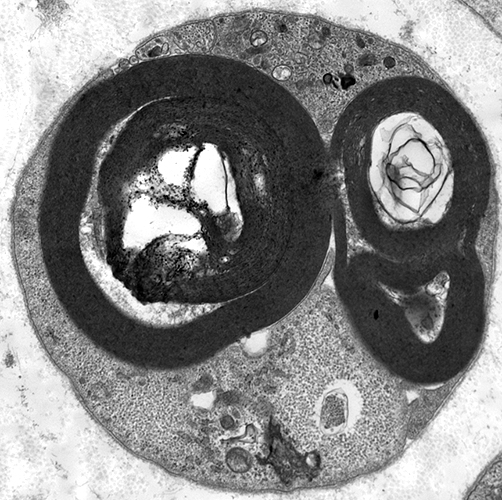 Electron micrograph: From Robert E Schmidt MD |
Myelin, compact: Fragmentation inside Schwann Cell
Schwann cell characteristic: Surrounded by Basal lamina (Below; Arrow)
Axon is lost
Autophagic Schwann cell
Contains: Myelin fragments & Lipid droplets
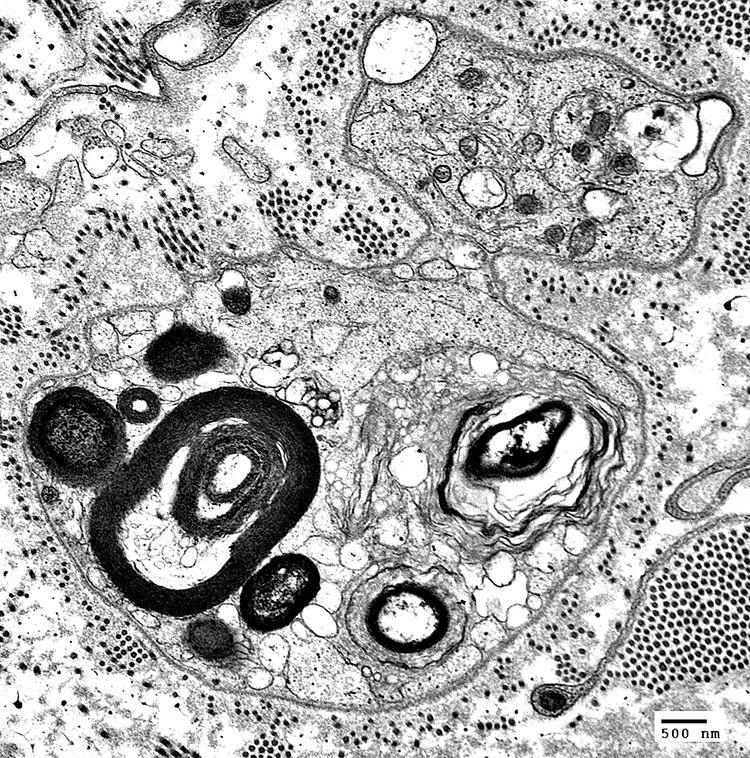
|
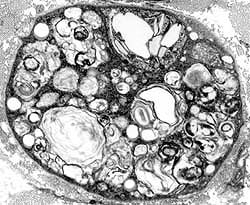 Robert E Schmidt MD Myelin Degradation: Later phase Autophagic Schwann Cells contain Lipid debris Myelin fragments, small Axons: Lost |
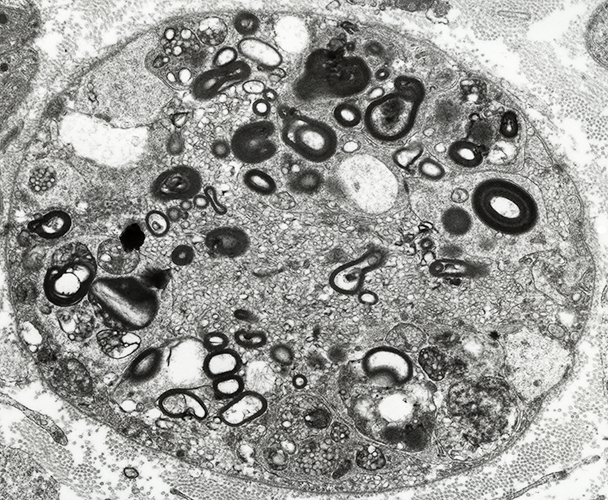 Electron micrograph: From Robert E Schmidt MD |
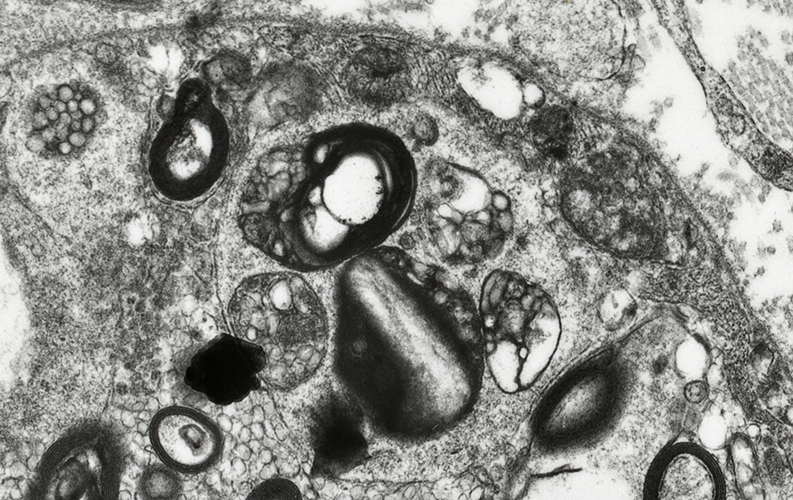 Electron micrograph: From Robert E Schmidt MD |
|
Myelin & Axon Degeneration: Ongoing, Weeks; Myelin Degradation to Lipids
| Ultrastructure |
Macrophages & Phagocytic cells 15
- Sources
- Intrinsic
- Recruitment from circulation
- Schwann cell differentiation
- Recruitment factors
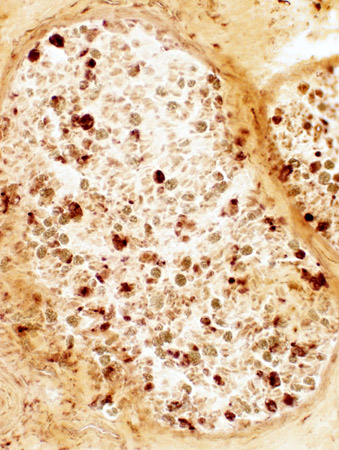 Acid phosphatase stain Autophagic/Phagocytic cells, Endoneurial Myelin sheaths are stained |
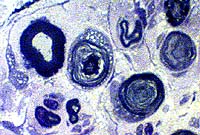 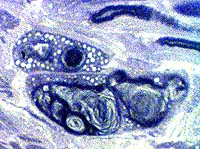 Toluidine blue stain Schwann cells contain Degraded Myelin sheaths Lipid droplets |
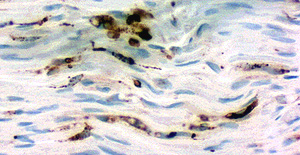 CD68 stain Macrophages/Autophagic cells |
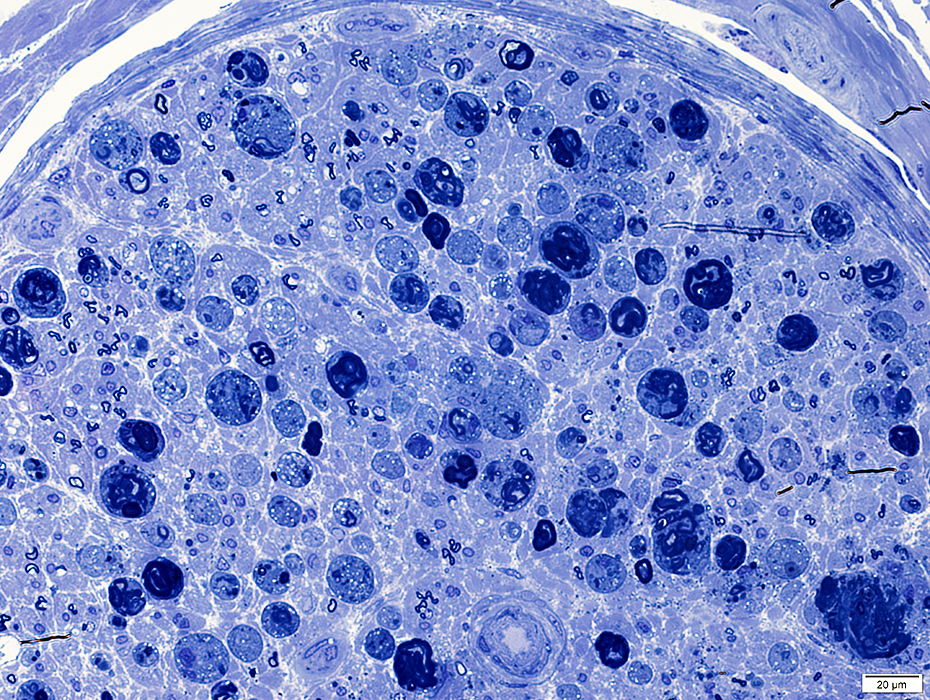 Toluidine blue stain |
Irregular, large myelin figures
Many histiocytes: Contain small round lipid droplets in cytoplasm
Axons are degraded & lost
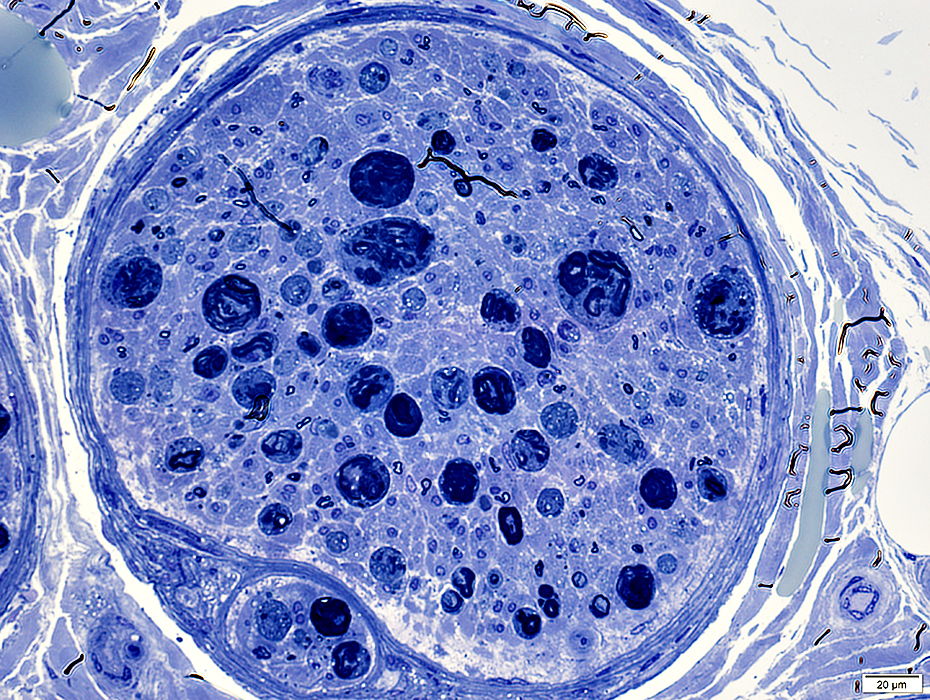 Toluidine blue stain |
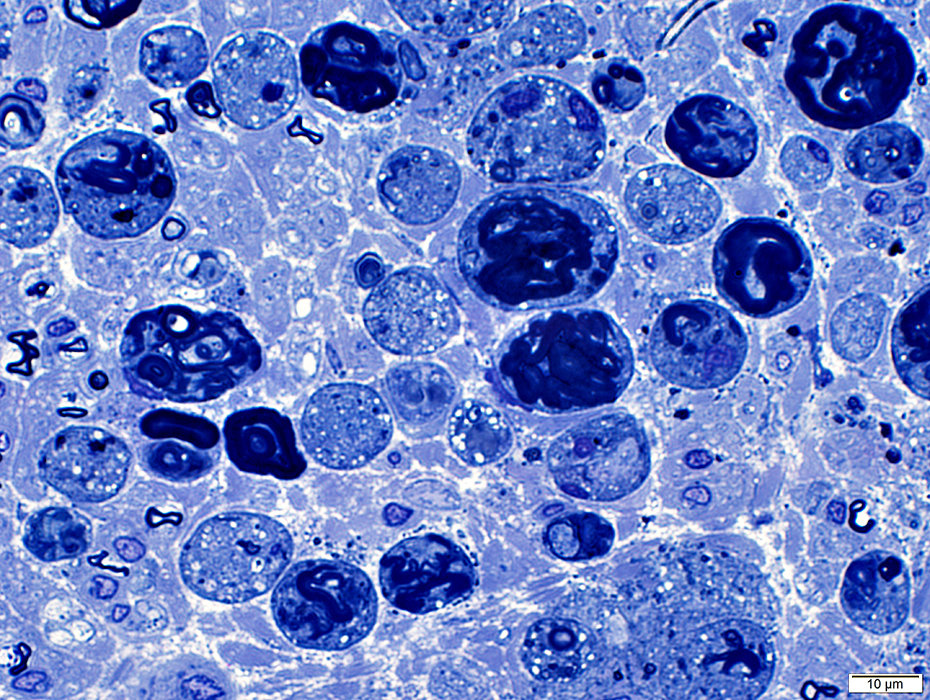 Toluidine blue stain |
Irregular, large myelin figures
Many histiocytes with lipid droplets in cytoplasm
Axons are degraded & lost
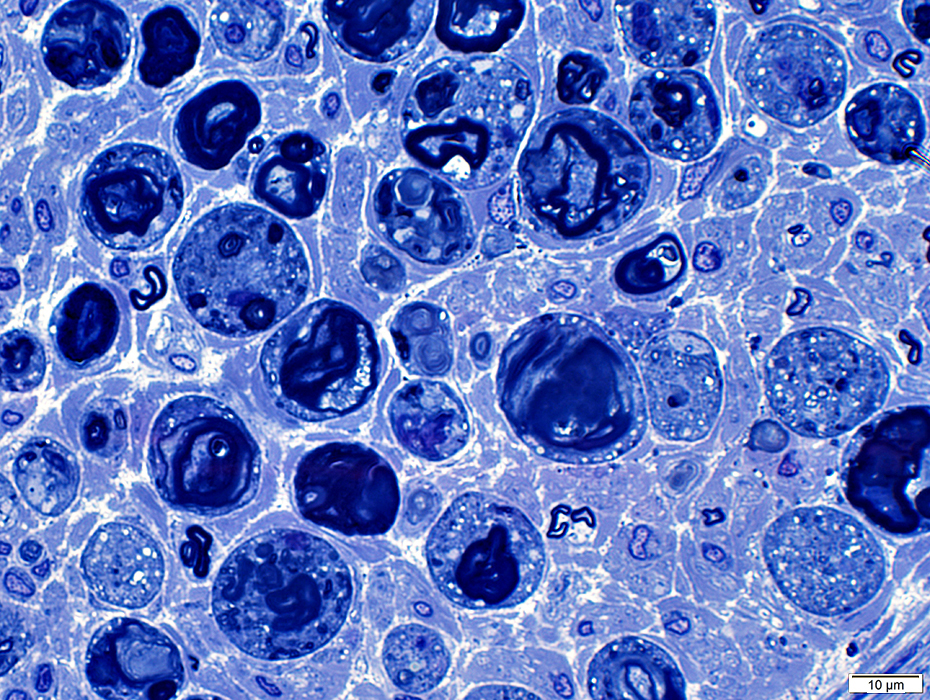 Toluidine blue stain |
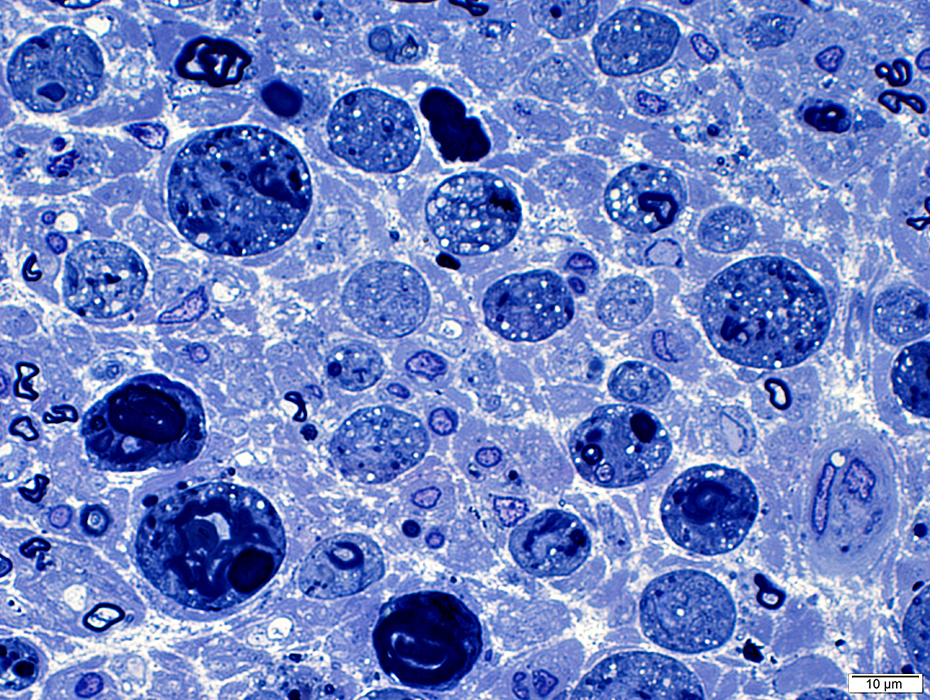 Toluidine blue stain |
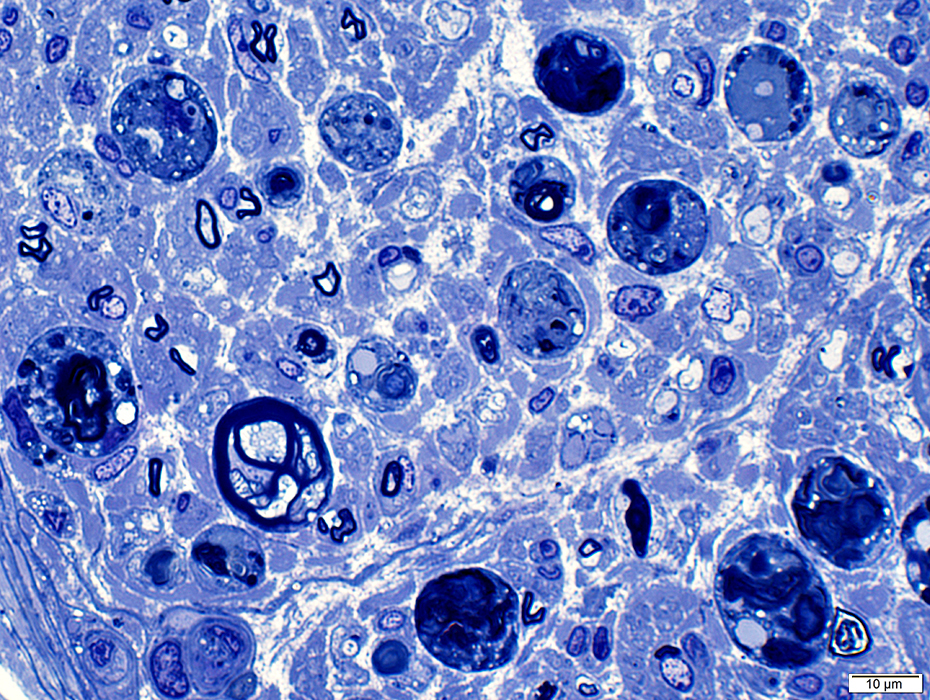 Toluidine blue stain |
Myelin Ovoids & Remnants
Myelin ovoids: Longitudinal section of nerve
 Toluidine blue stain |
Myelin Ovoids: Teased axons
Top: Myelinated axon, control (Node of Ranvier at Arrow)
2nd row: Schwann cell sheath with no remaining myelin fragments or axon
Below: Myelin ovoids & remnants along paths of previously degenerated myelinated axon
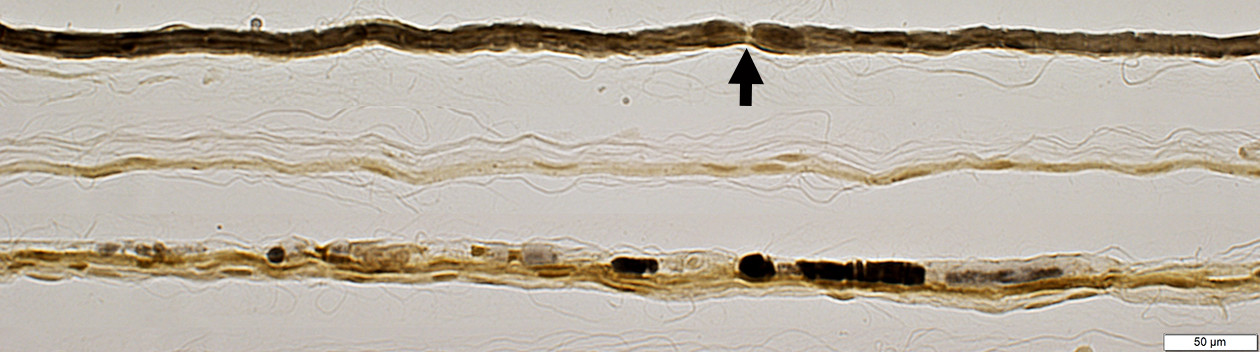
|

|
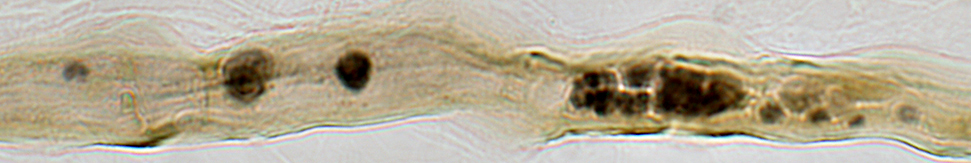
|
Myelin Remnants: Features
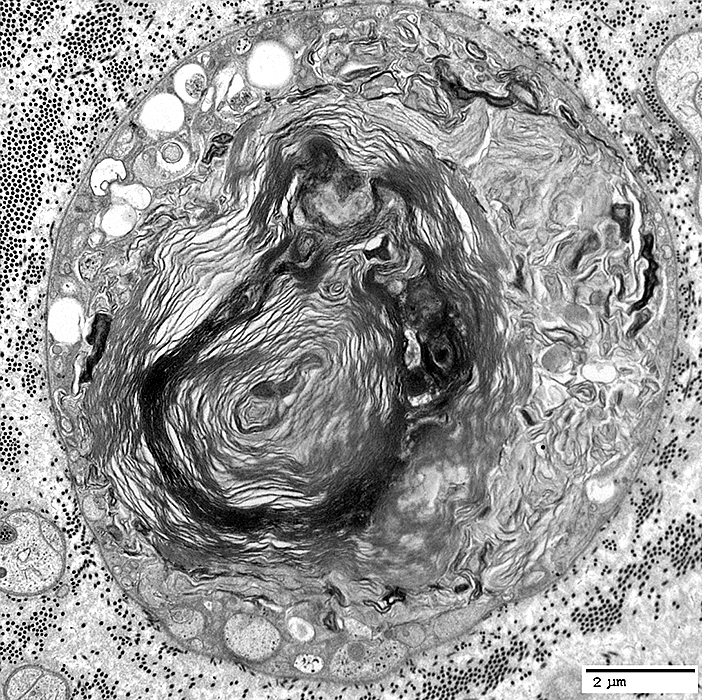
|
Within Schwann cell
No associated axon
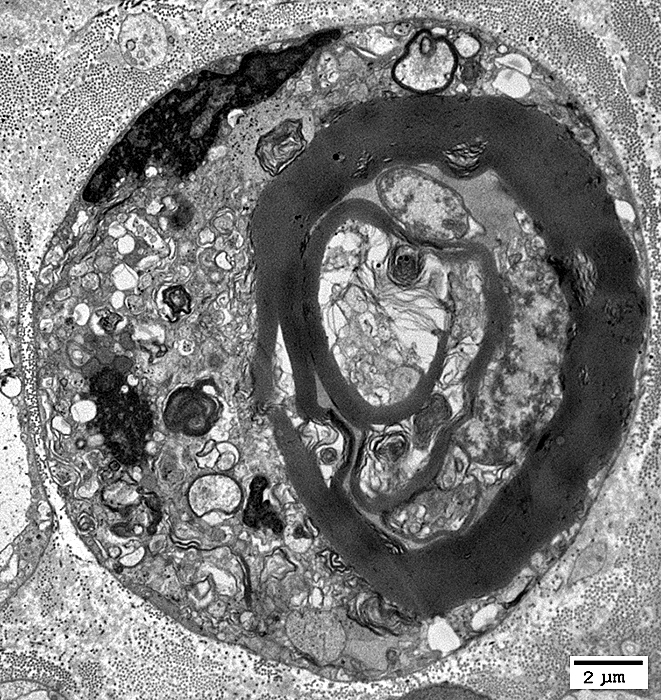
|
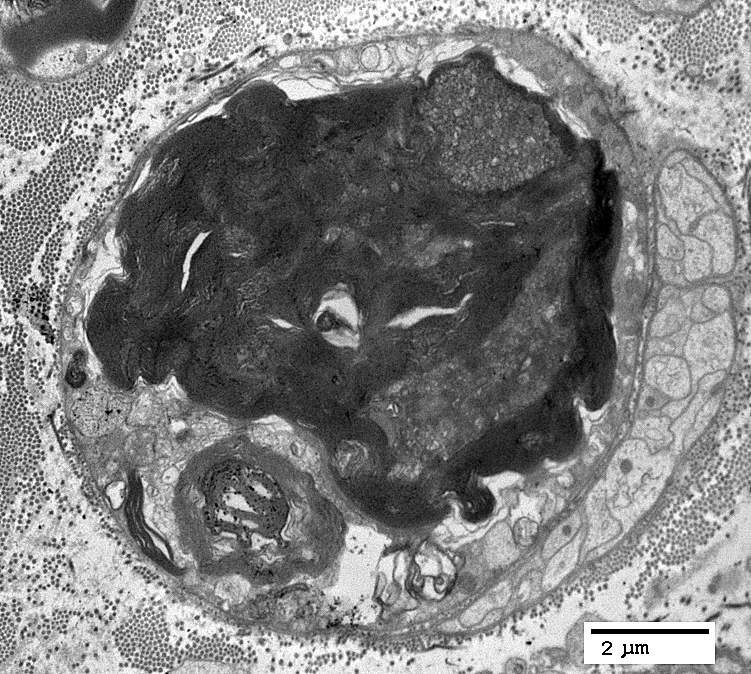
|
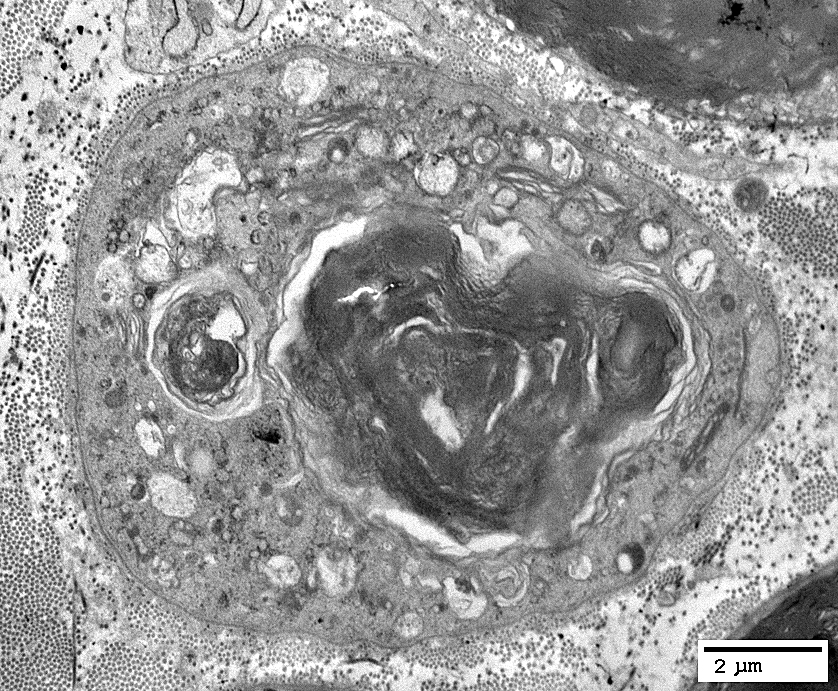
|
Histiocytes: Debris- & Lipid-containing
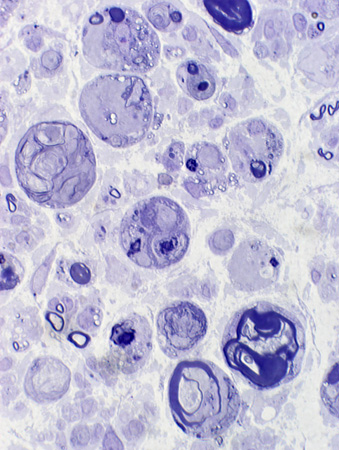
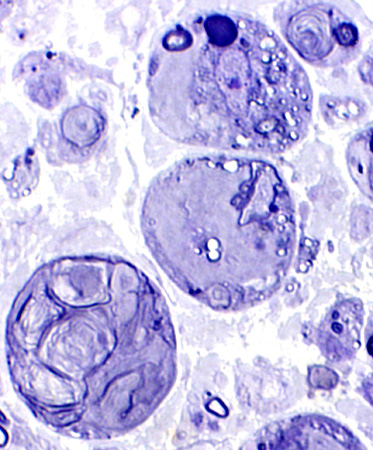
|
Large cells: Contain myelin debris or round, clear lipid droplets.
Normal axons with thin & thick myelin sheaths may also be present.
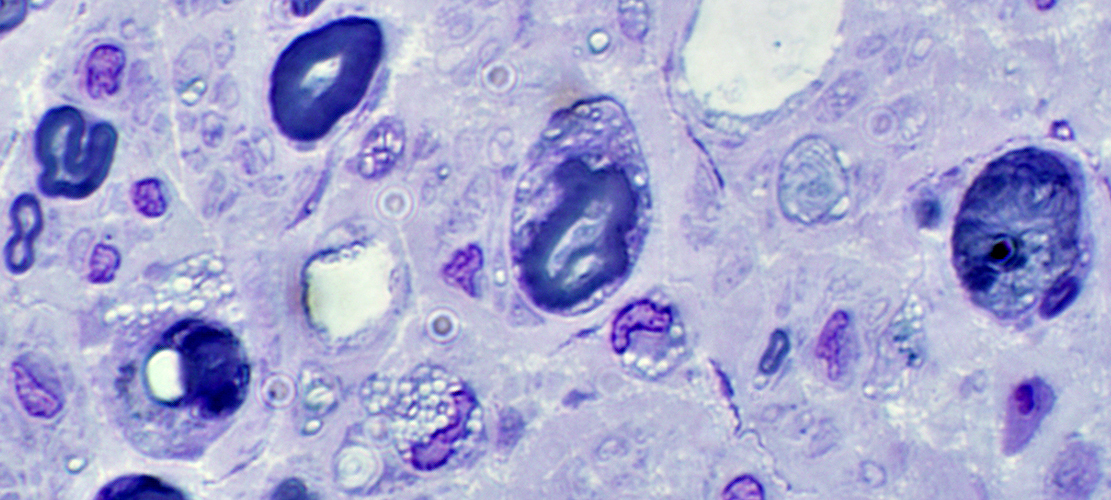
|
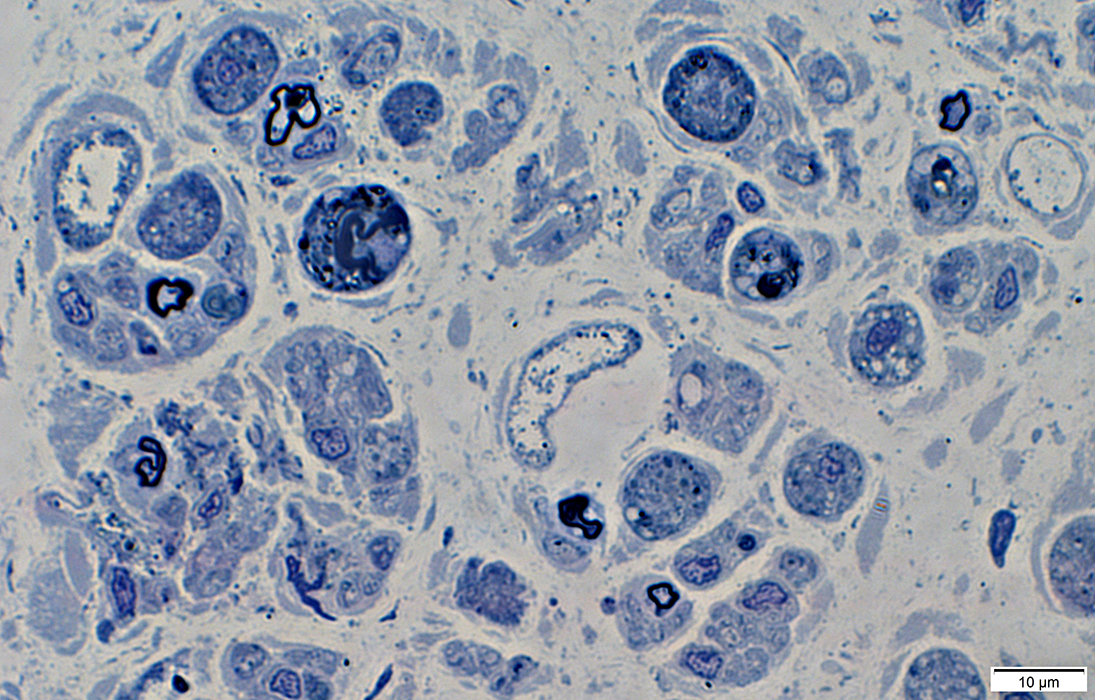 Toluidine blue stained plastic sections |
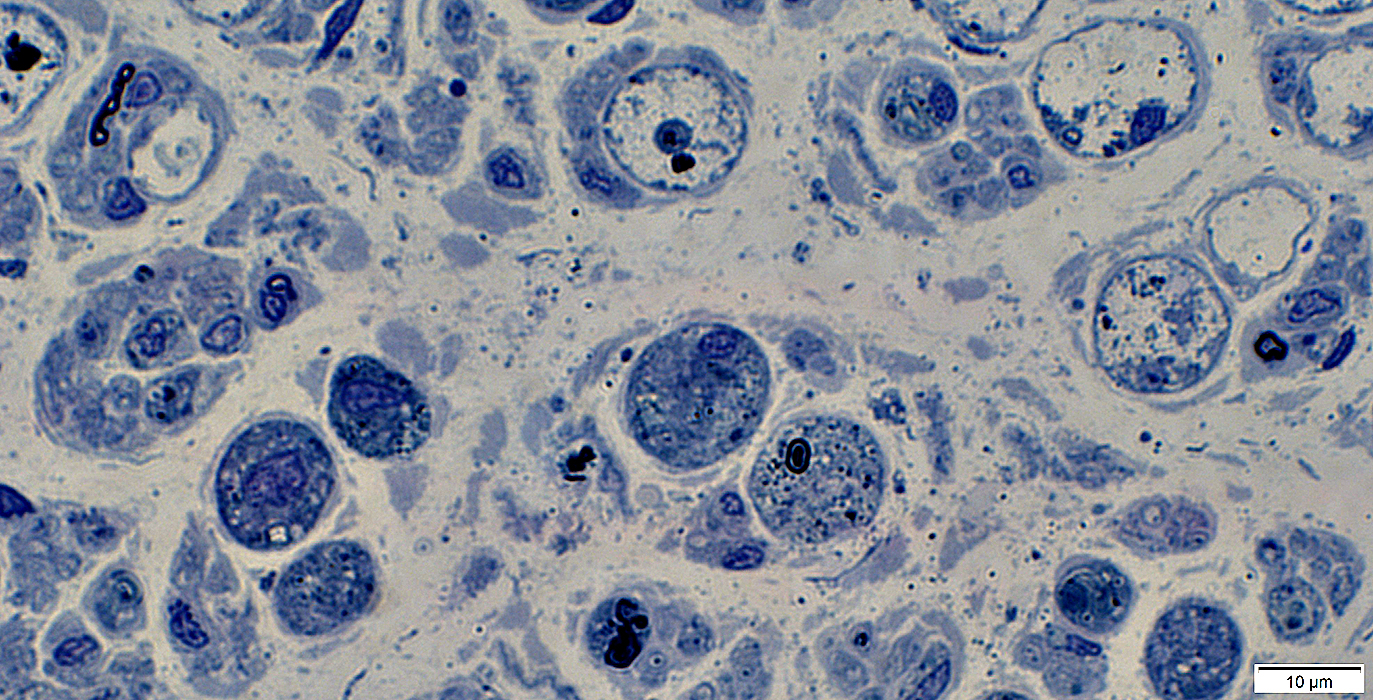 Toluidine blue stained plastic sections |
Early WD: Histiocytes
Myelin
Myelin outer layers: Irregular structure
Schwann cell, Ab-Axonal cytoplasm
Contains myelin fragments
Histiocytes
Present in areas around axon
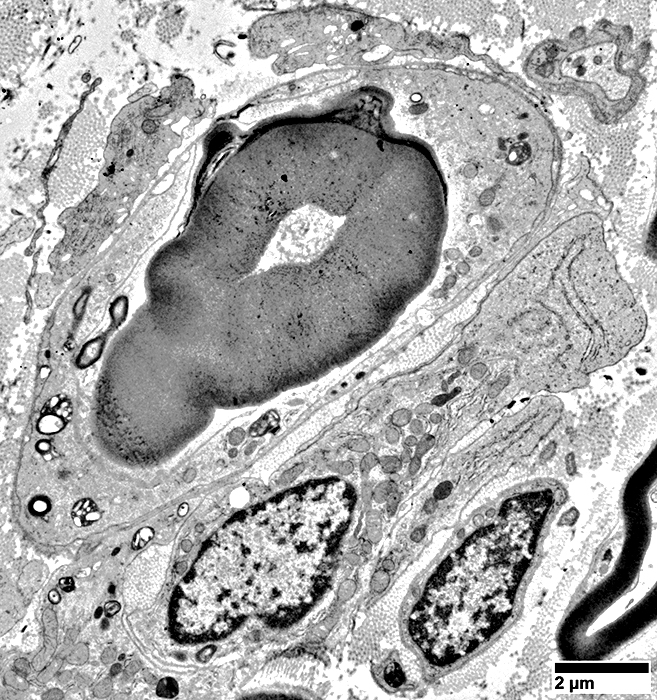 |
Histiocytes with Myelin fragments around a cell with a large, partially degraded Myelin sheath
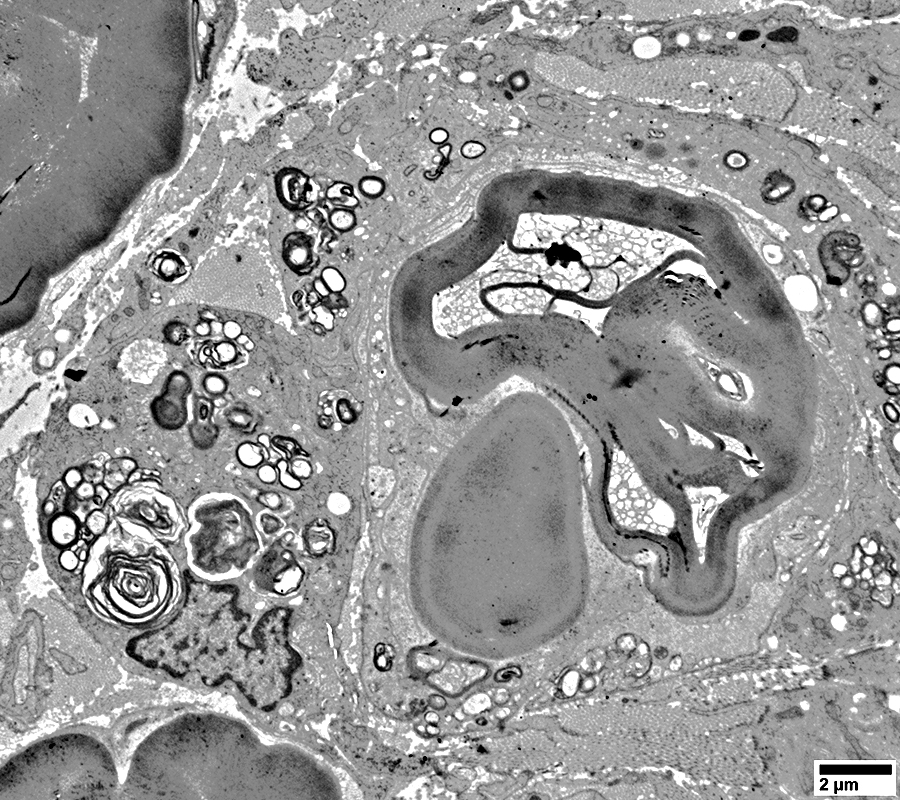
Myelin Degradation: Late stage with Lipid droplets & some Myelin fragments
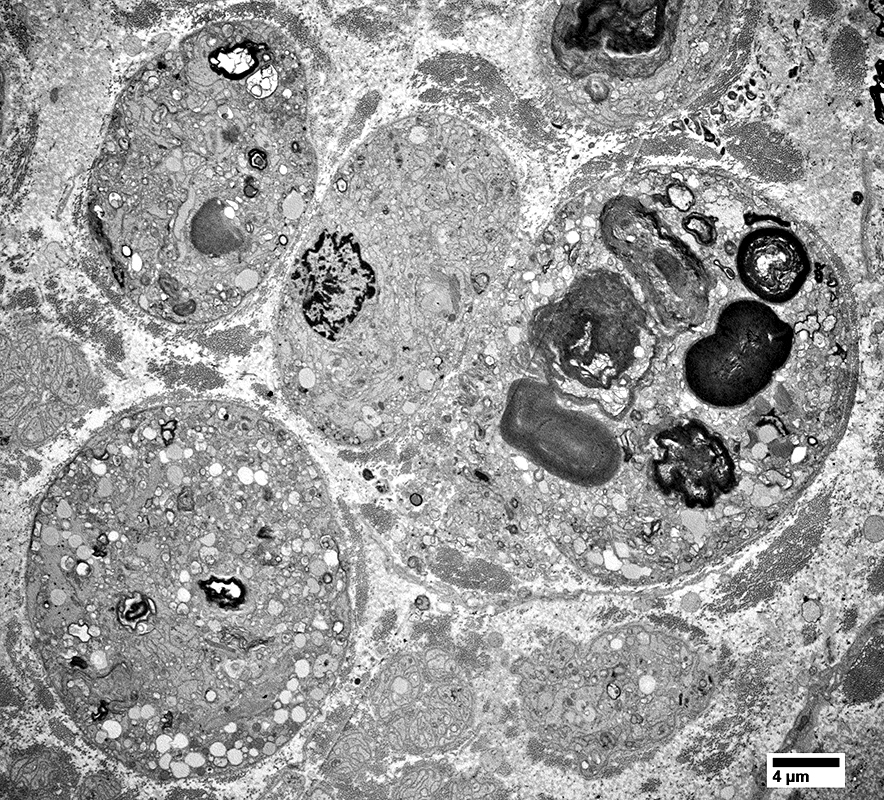
|
Some cells have mostly myelin debris (Upper right)
Some cells have many clear, round lipid droplets
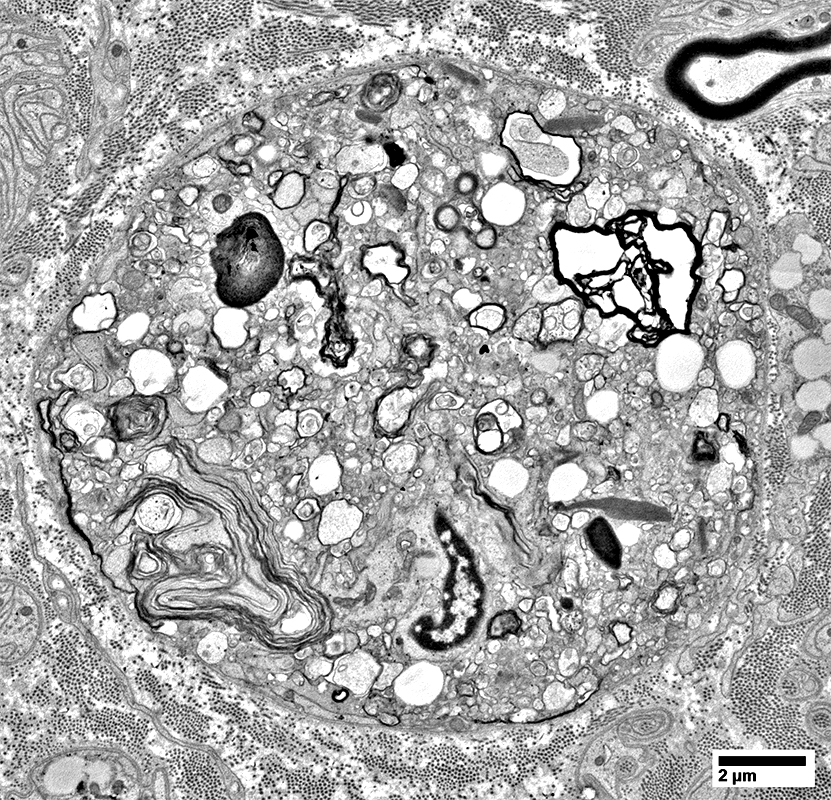
|
Schwann cell processes: Surrrounded by a single basal lamina
Some contain myelin in different stages of degradation
Degradation products include
Ovoid-like structure (Right)
Lipid droplets of different sizes
Some smaller Schwann cell processes have no degradation products

|
Histiocyte: Endoneurial
Contains many clear, round Lipid droplets
Surface: Shows several processes extending into endoneurium
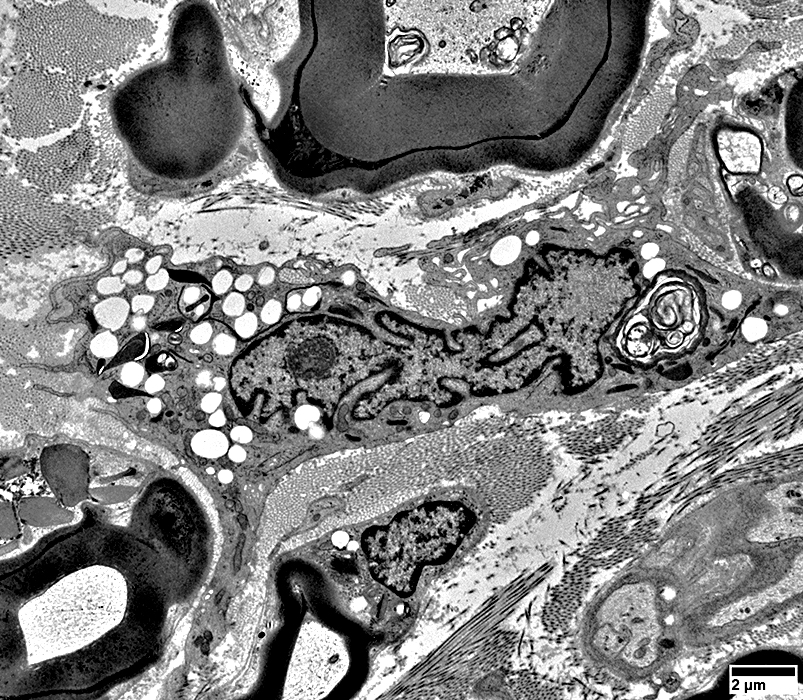
|
Lipid droplets in Schwann Cells & Macrophages
|
Schwann cells (Dark arrows) & Macrophages (Light arrows) Closely-apposed Phagocytic Contain lipid droplets & Myelin debris |
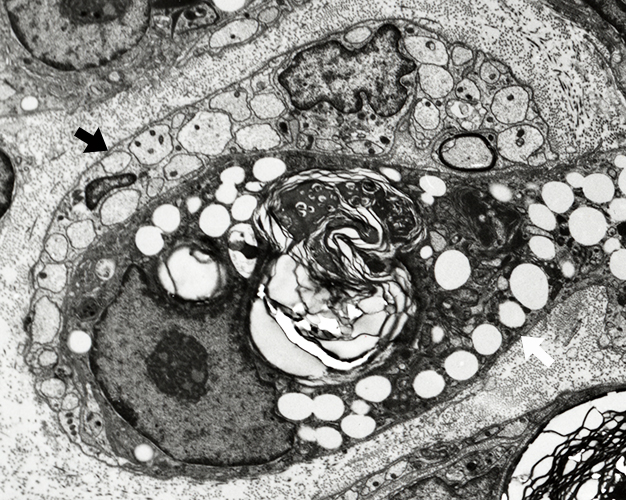 |
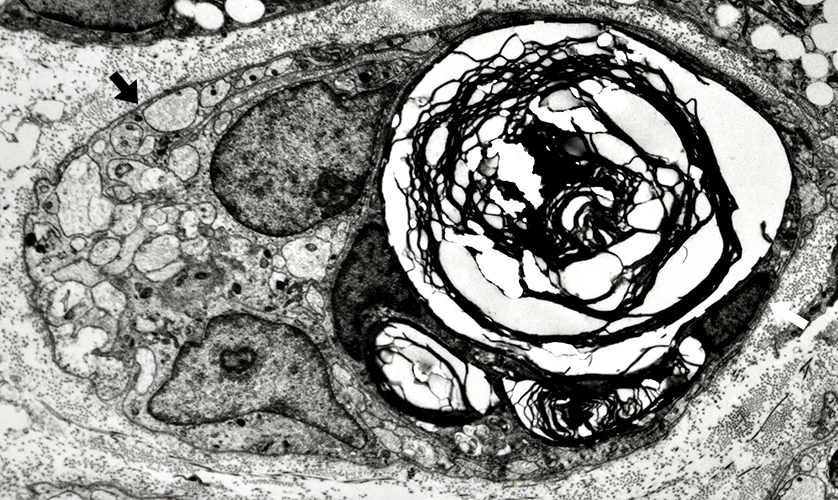 |
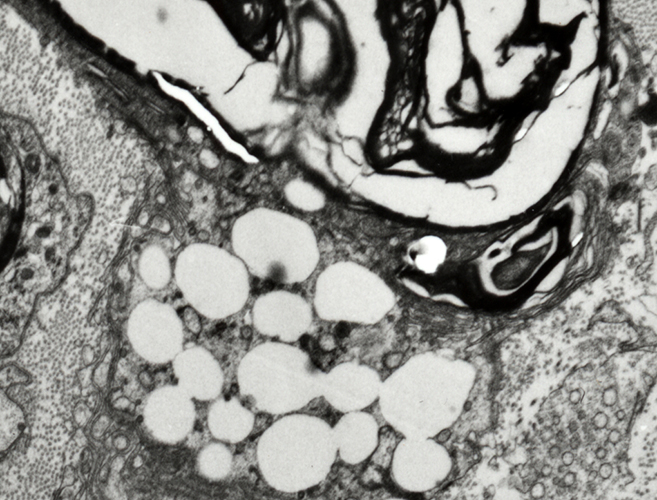
|
Wallerian Degeneration, Later stage: Histiocytes with Lipid debris around Endoneurial Microvessels
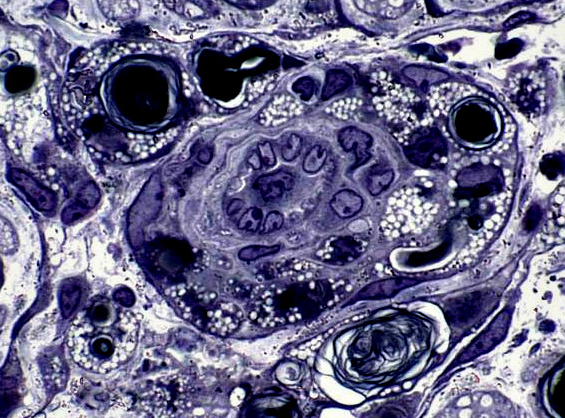 From R E Schmidt MD |
- Contents
- Lipid droplets
- Myelin debris, small
- Location
- In & around vessel wall
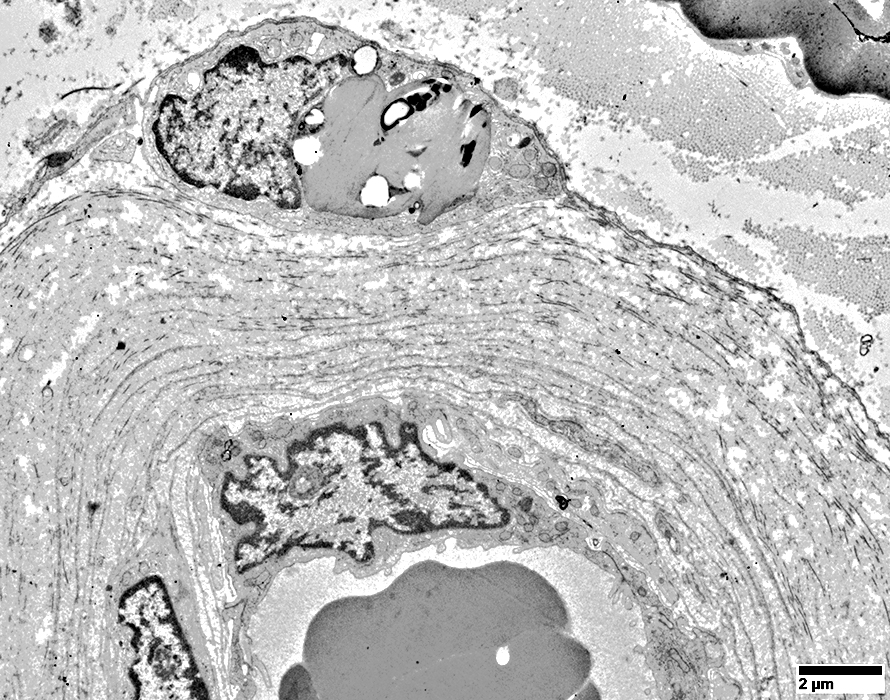 From R E Schmidt MD |
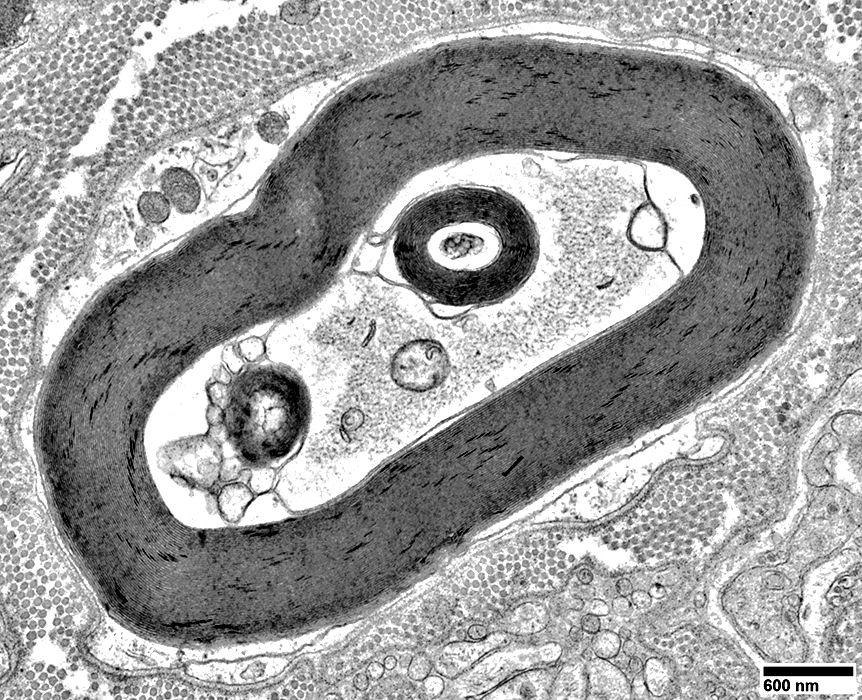
Axons, Regenerating/RegeneratedSurrounded by Schwann cell cytoplasm Unmyelinated (Below) Thinly myelinated axon (Far right) 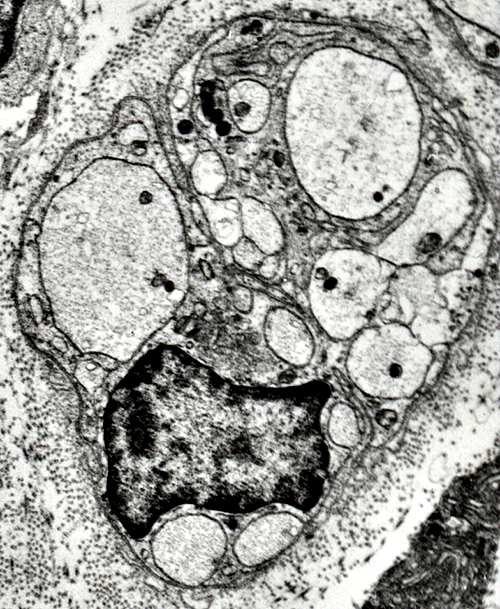 From R E Schmidt MD |
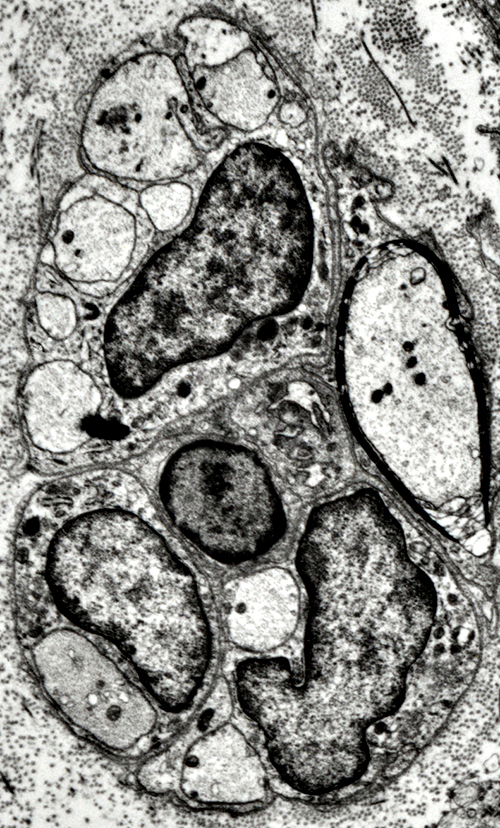 From R E Schmidt MD |
Axon Sprouts & Growth Cones
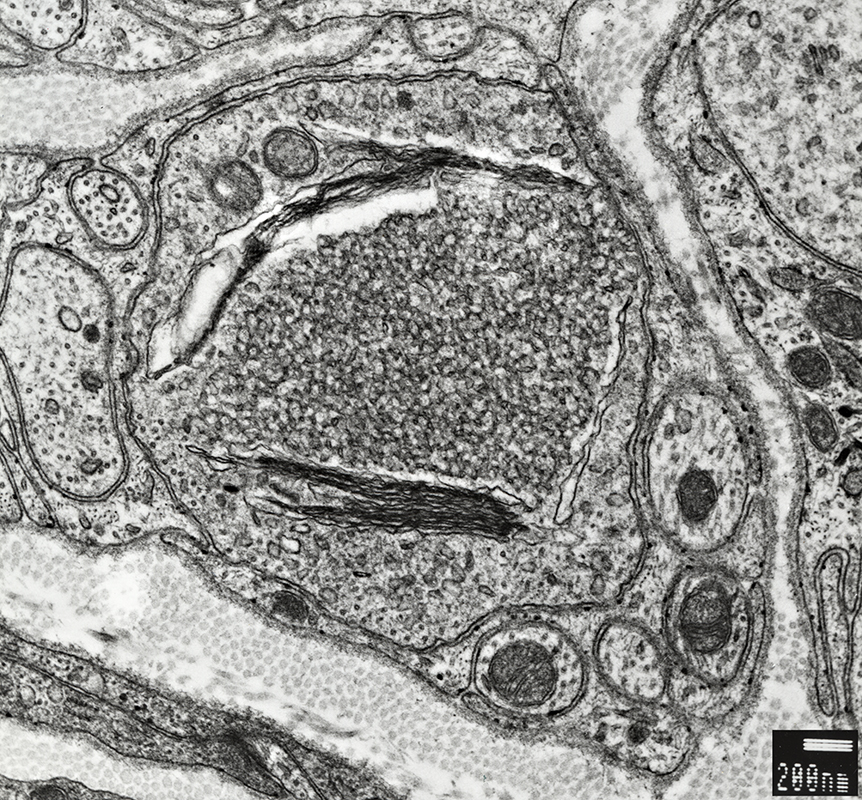 From R E Schmidt MD |
Contains: Tubulovesicular material
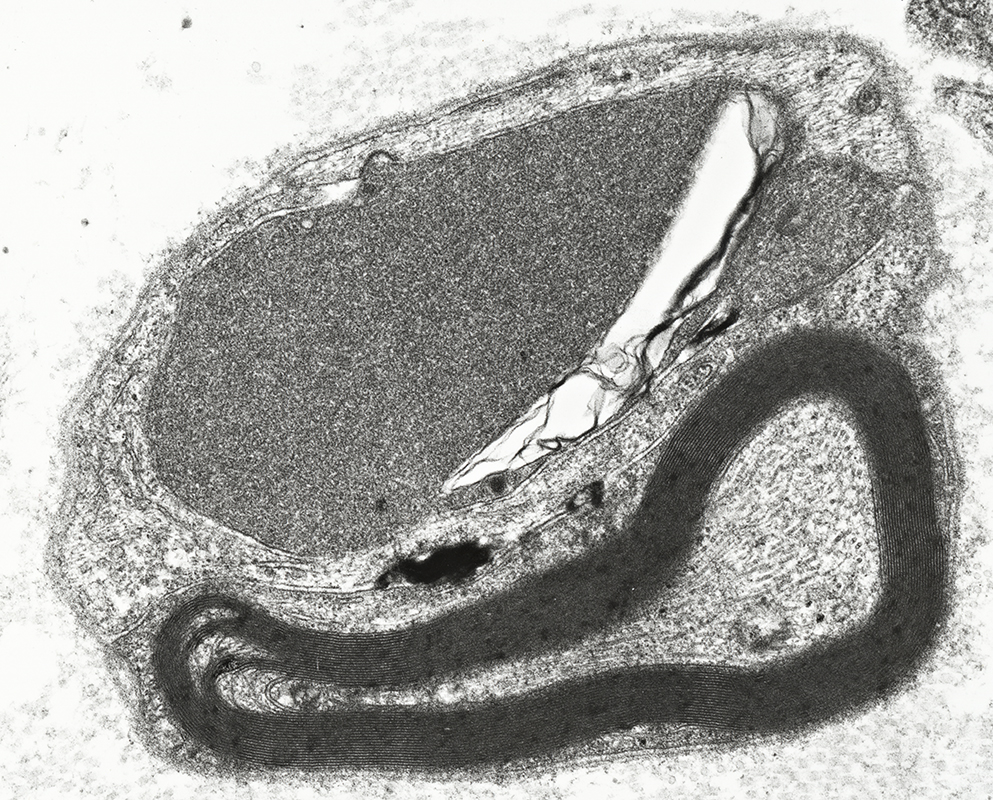 From R E Schmidt MD |
Contain: Mostly filaments
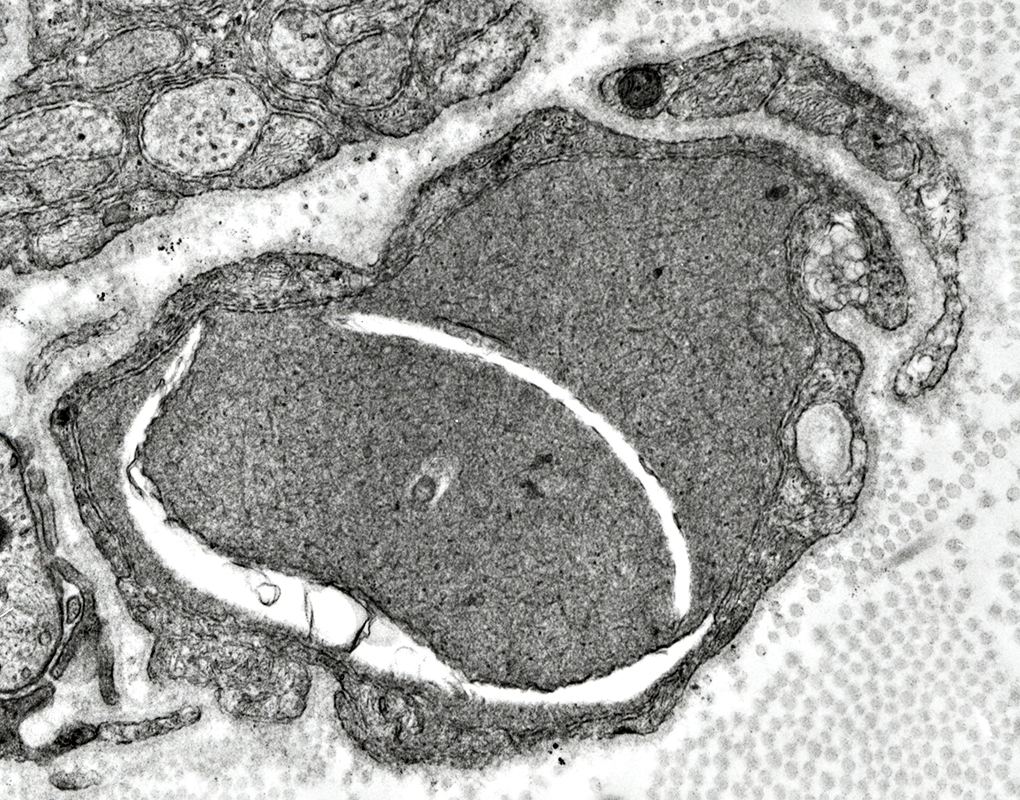 From R E Schmidt MD |
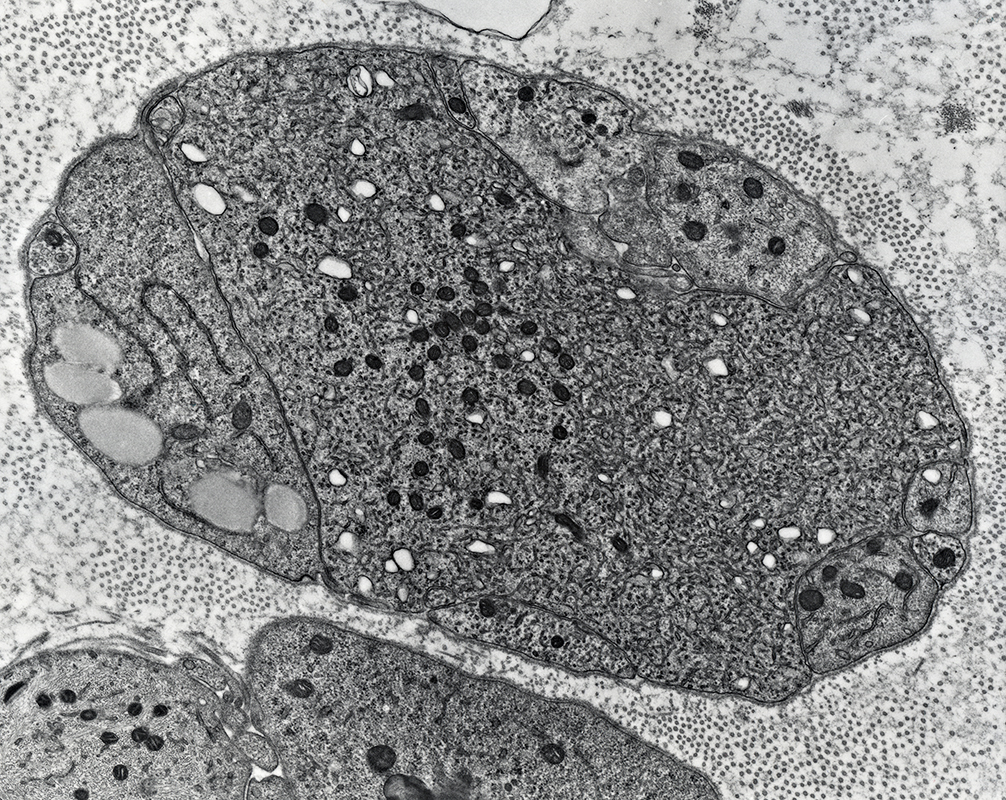 From R E Schmidt MD |
Contains: Vesicular material & mitochondria
Surrounded by: Schwann cell processes & basal lamina
|
Denervated Schwann cell bands (Bands of Büngner)
|
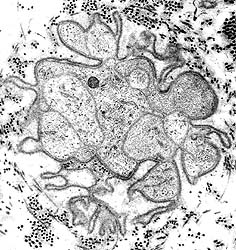 From R E Schmidt MD |
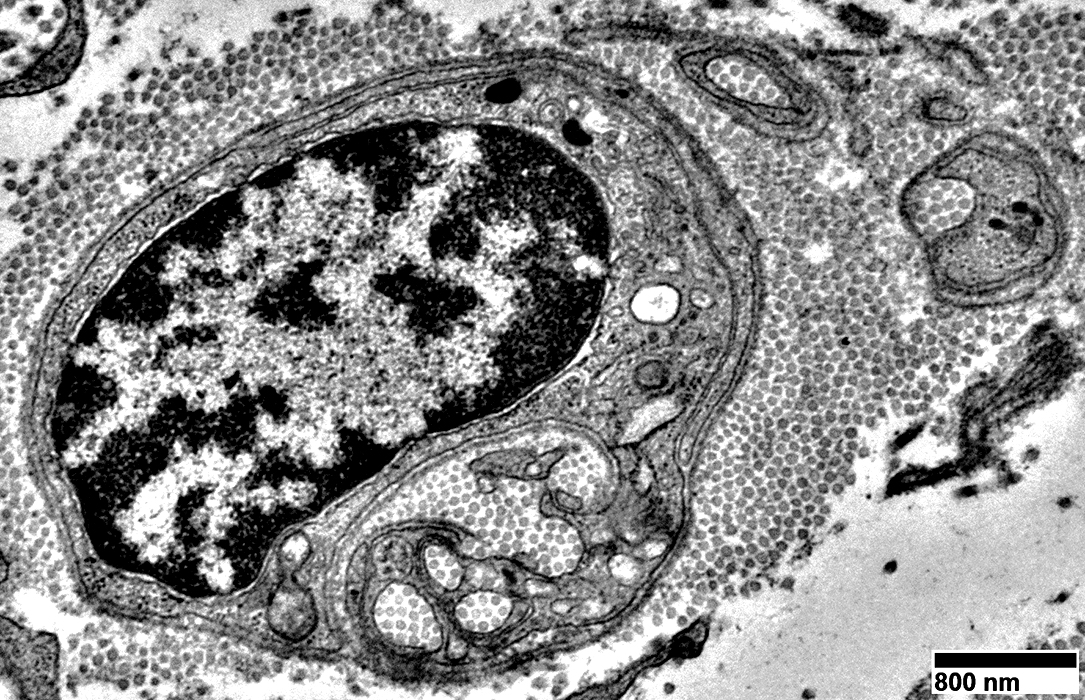
|
Collagen PocketsCollagen fibrils surrounded by Schwann cell processesMore common with Loss of small axons Increased age No axon regeneration 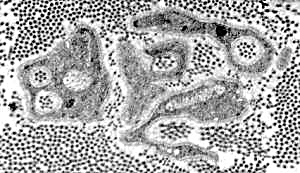 RE Schmidt MD |
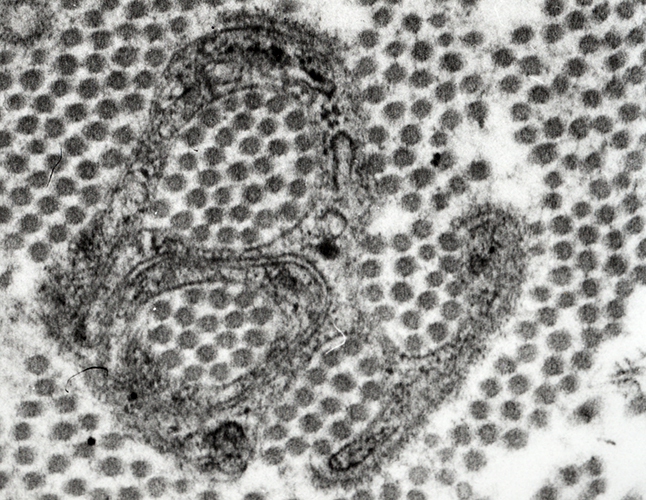 Electron micrograph: From Robert E Schmidt MD |
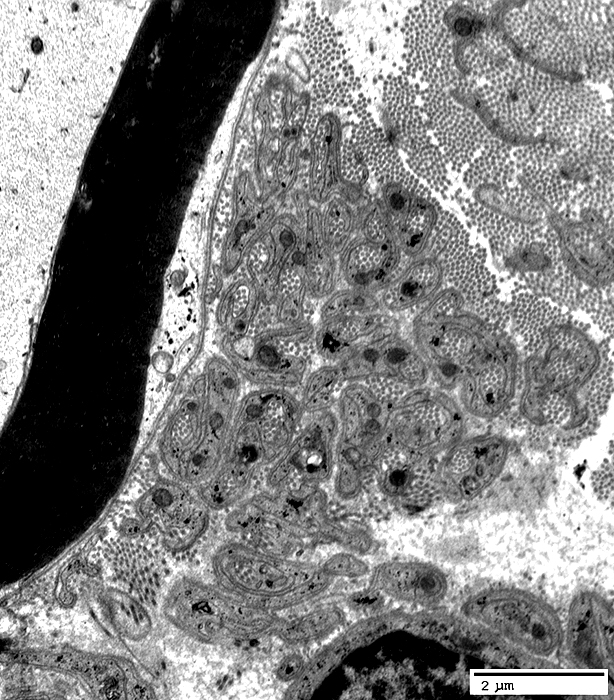
|
Collagen fibrils surrounded by Schwann cell processes
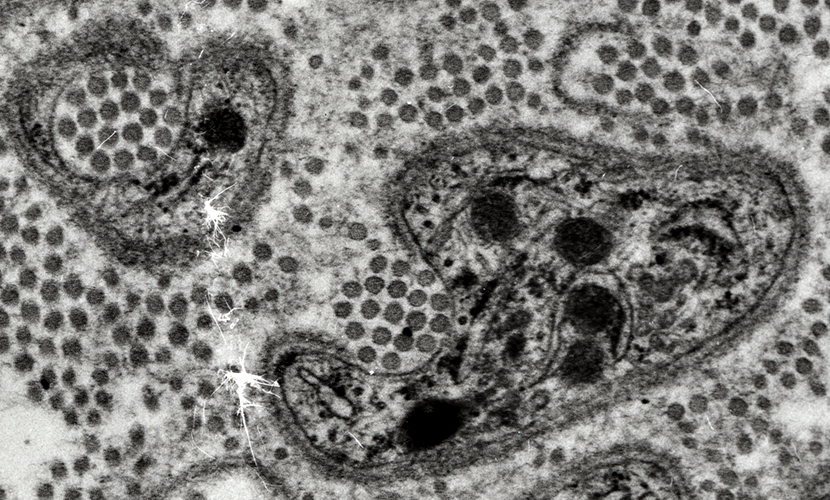 Electron micrograph: From Robert E Schmidt MD |
Schwann cell processes within & surrounding collagen fibrils
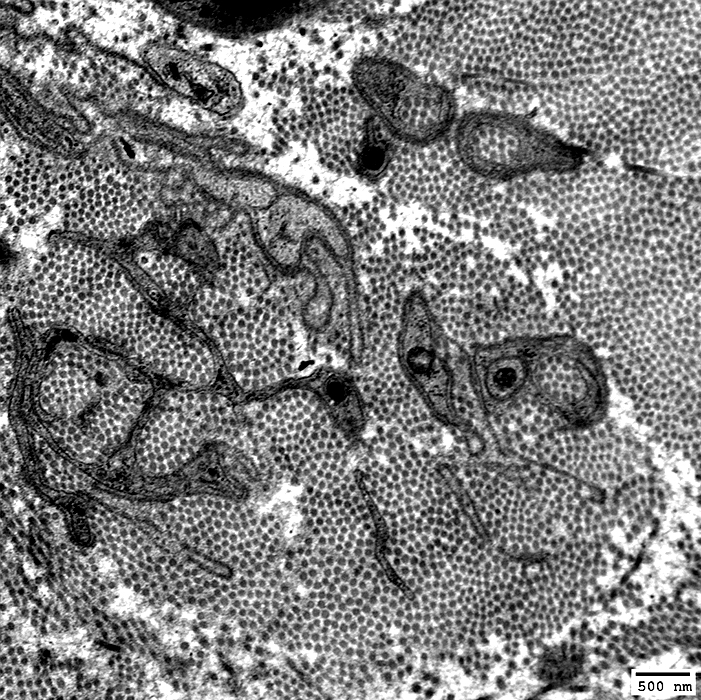
|

|
 From: R Schmidt |
Collagen Pocket: Large
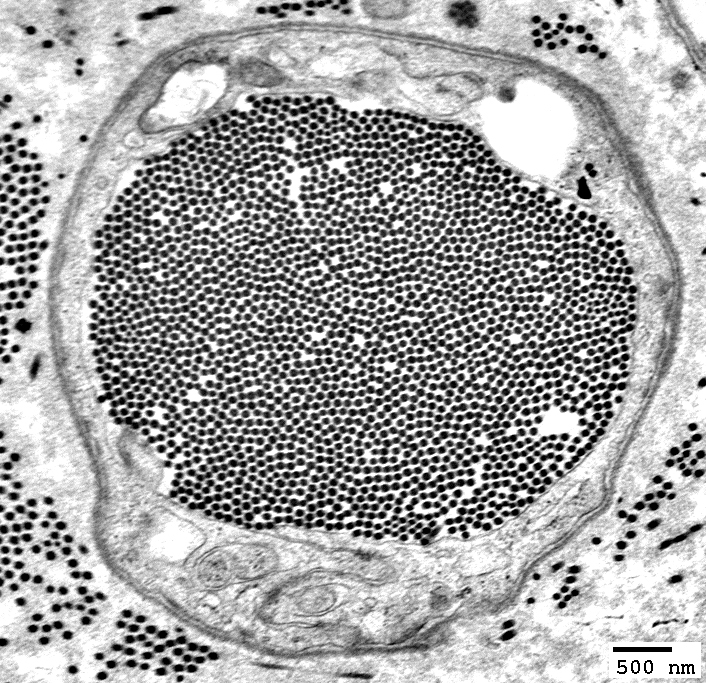 From: R Schmidt |
Loss of Myelinated, & Some Unmyelinated, Axons
Unmyelinated axons: Mild lossNormal: Yellow (Green axon + Red NCAM stained Schwann cell)
Lost: Non-myelinating Schwann cells (NCAM stained, Red) without associated axons (Arrow)
Myelinated axons: Severe loss
Markedly reduced numbers of larger green (neurofilament) axons
Few remaining large axons are small
See
More severe small axon loss
Control (Below)
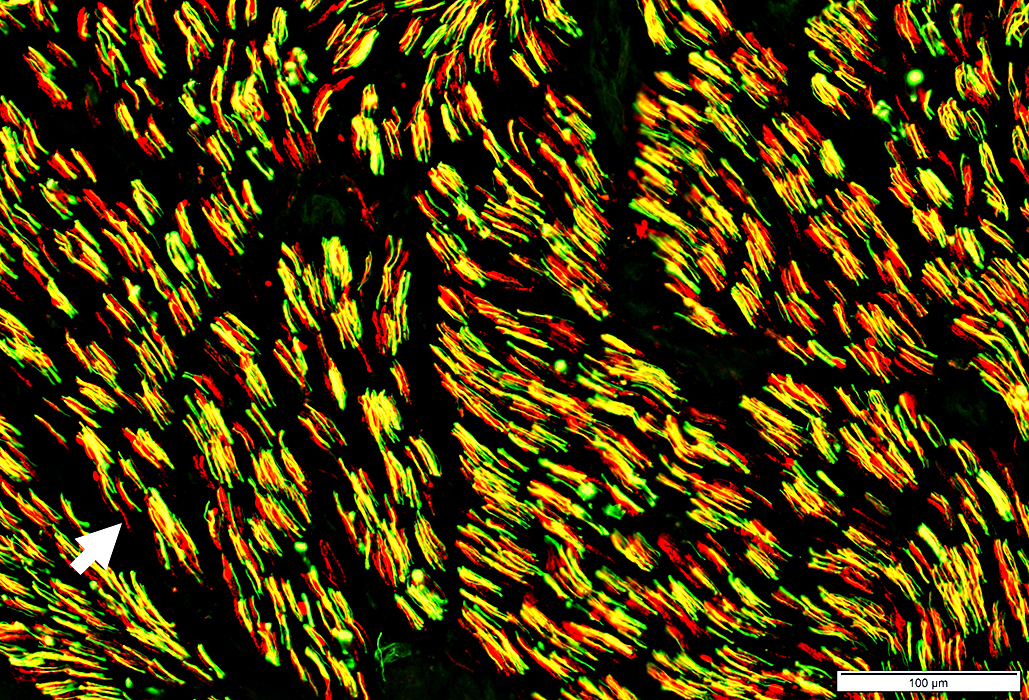 Neurofilament stain (Green) + NCAM stain (Red) |
Large Axons: Nearly complete loss (Chronic)
Large axons: None stained
Small axons: Many preserved
Myelin basic protein: Co-stains (Yellow) on many smaller axons
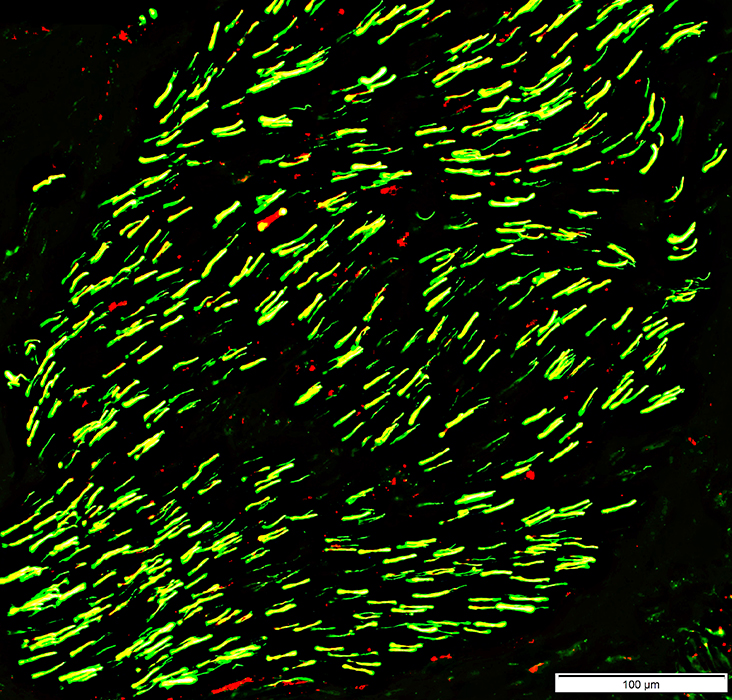 Neurofilaments (Green); MBP (Red) |
Control nerve
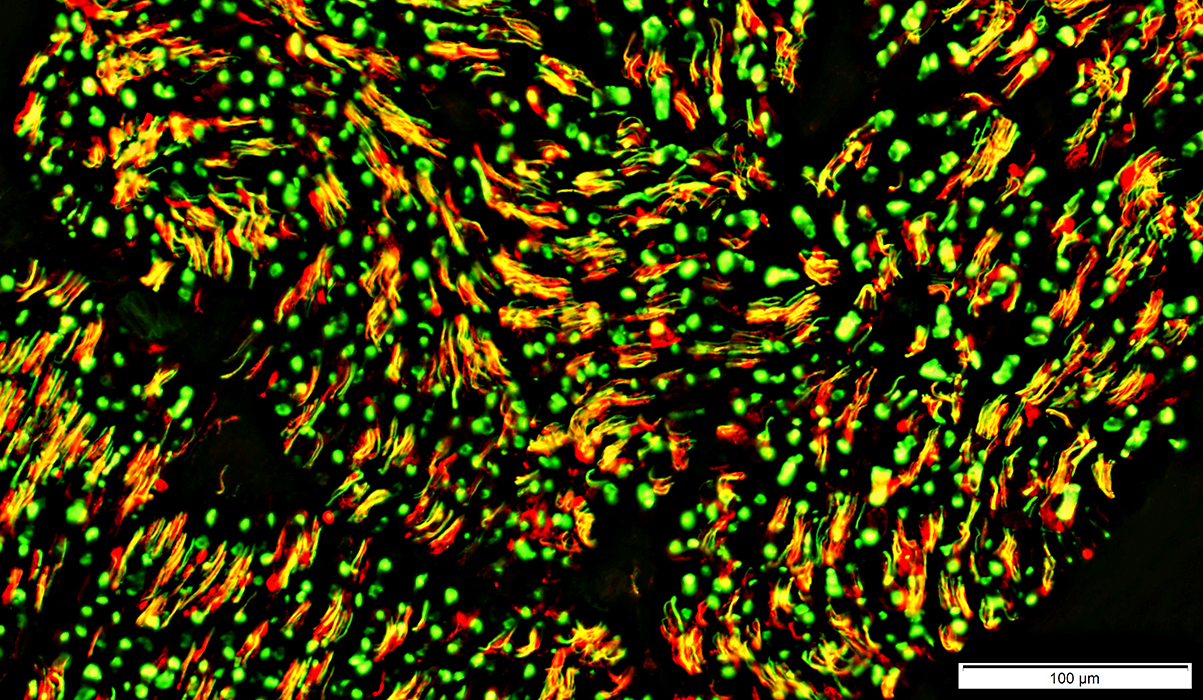 Neurofilaments (Green); NCAM (Red) |
2 sizes: Large & Intermediate
No associated NCAM positive (red or yellow) cells
Unmyelinated axons (Small, Yellow)
Occur in clusters
Co-stain (Yellow) for neurofilaments & surrounding non-myelinating Schwann cells
Also see
Large axon loss
Large & Small axon loss
Skin: Normal & Axon loss
|
Biopsy General Images Technical features |
Skin Innervation & Biopsy: General
- Utility: Reliable test to document loss of small axons in skin
- Measurement: Intraepidermal nerve fiber density (IENFD)
- Patterns of axon loss & damage
- Length dependent
- Definition: More loss of axons in distal than proximal leg
- Symptoms: Distal; Symmetric
- Associations
- Disorders that damage axons
- Diabetes: More common than in Non-length dependent small fiber neuropathies
- Non-length dependent
- Definition: Similar, or more, loss of axons at proximal compared to distal locations
- Association: Small fiber ganglionopathy
- Symptoms
- Involvement of face, mouth, trunk, upper limbs, or muscle
- Disease associations
- IgM antibodies vs TS-HDS
- IgG antibodies vs FGFR3
- Acute onset small fiber neuropathies
- Idiopathic
- Fibromyalgia-like syndromes
- Systemic immune disorders: Sjögren syndrome
- Axon beading: May be early sign of axon damage
- Painful neuropathies
- May have reduced miR-146a & miR-155 expression in painful regions
- Normal numbers of small axons
- May occur in: Hereditary pain syndromes
- Length dependent
- Autonomic C-fibers
- Vasomotor: Blood vessel walls
- Sudomotor: Sweat glands
- Pilomotor: Arrector pilorum smooth muscle
Skin: Normal innervation
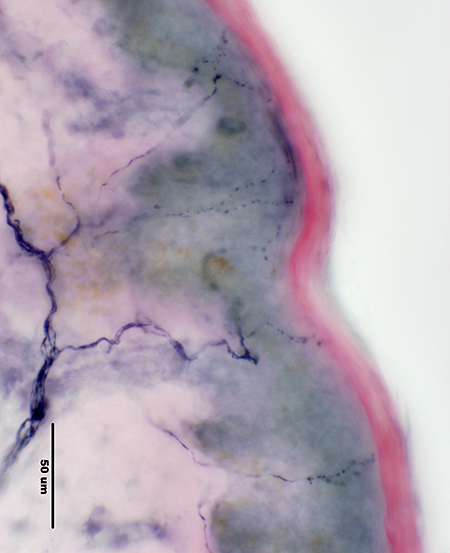 PGP 9.5 stain: Glenn Lopate |
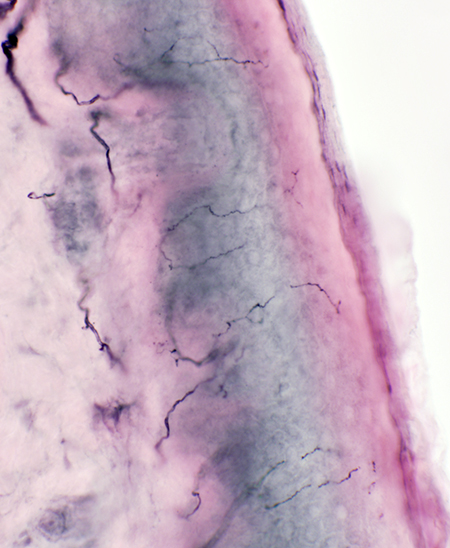 |
Skin: Pathologic innervation
Beaded axons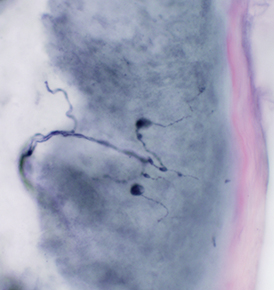 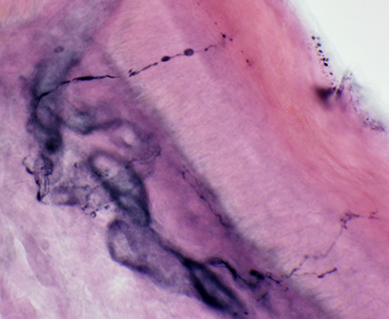 PGP 9.5 stain: Glenn Lopate |
Axon loss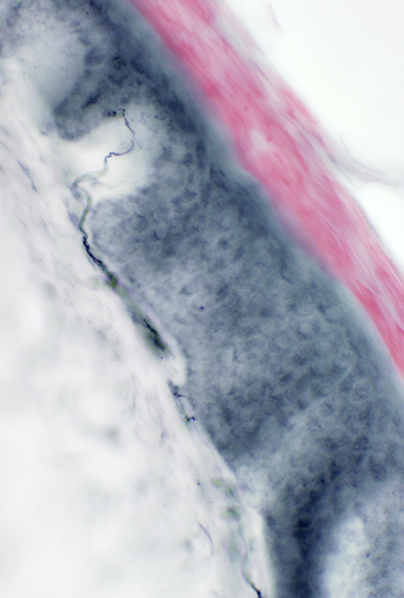 |
Myelin Artefact
Vesiculated myelin
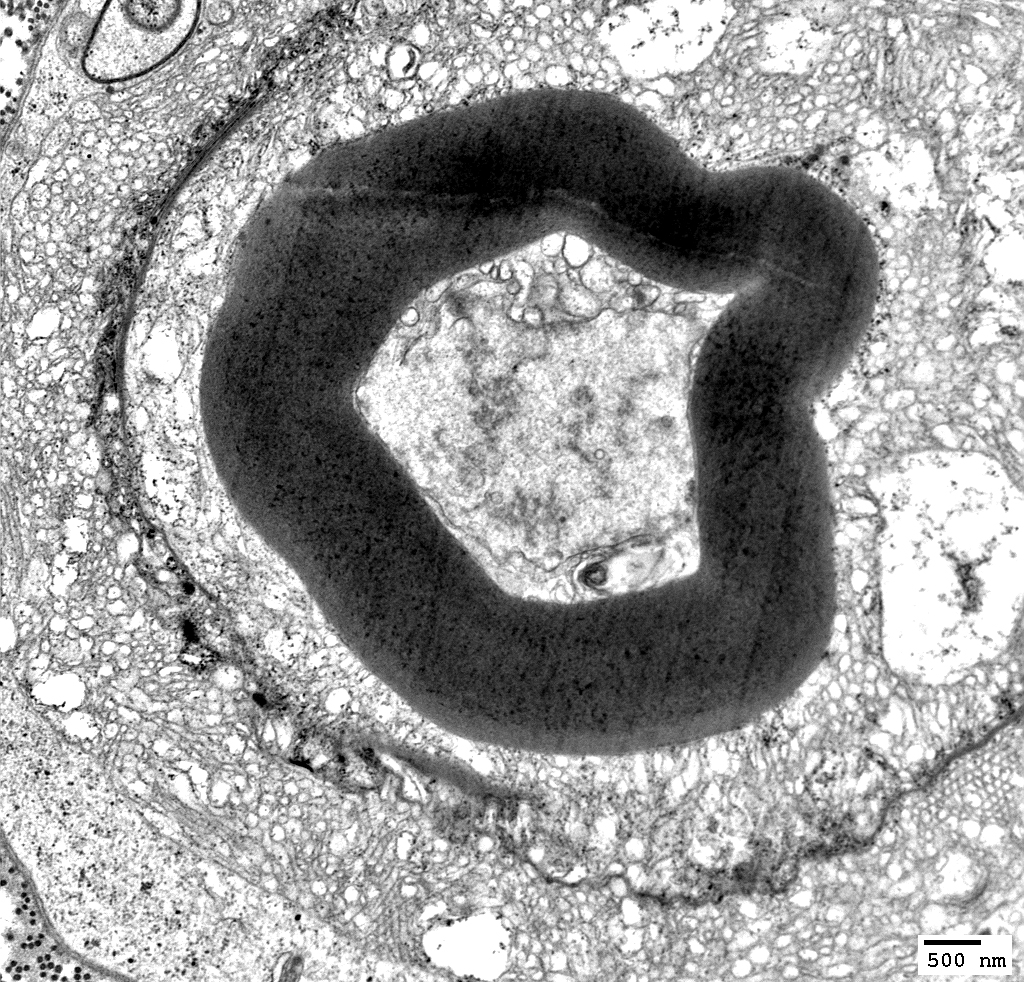
|
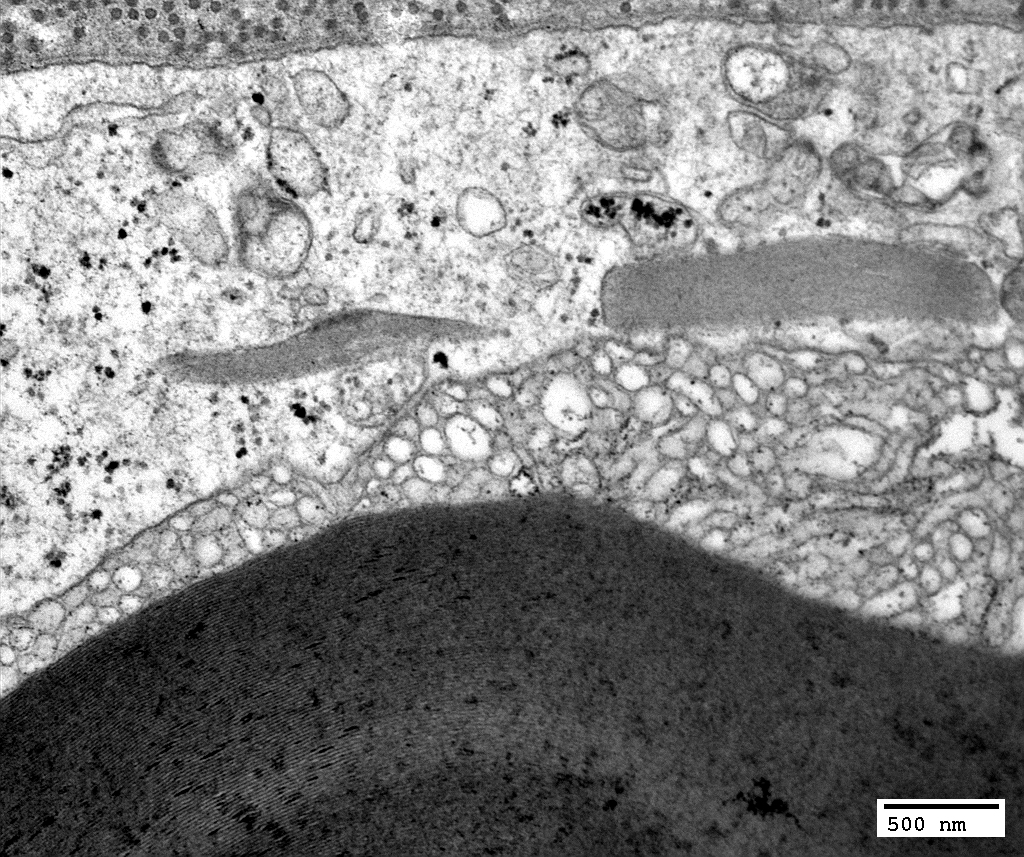
|
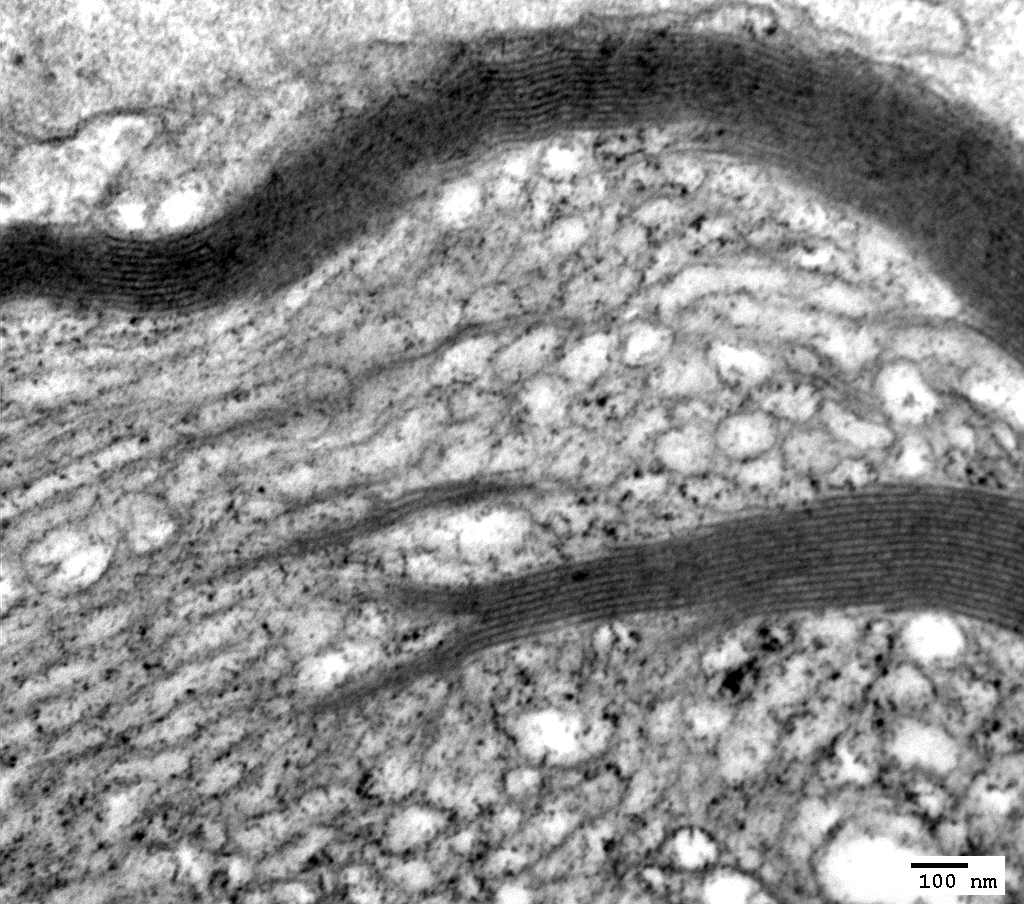
|
Go to Normal nerve
Return to Pathology & Illustrations
Return to Neuromuscular Home Page
1. Curr Opin Neurol 2001;14:635-639, Science 2002;296:868-871
2. J Neurosci Res 2002;68:432-441
3. Neurobiol Dis 2005;19:293-300
4. J Neurosci 2005;25:3478-3487
5. J Cell Biol 2012;196;7–18, Annu Rev Genet 2021;55:93-113
6. J Cell Biol 2015; July
7. Neuron 2016;89:449-460
8. Neuron 2017;93:1334-1343.e5, Neuron 2021;109:1067-1069, Curr Opin Neurobiol 2024:87:102884
9. Curr Biol 2017;27:890-896
10. Glia 2018 Oct 30
11. Mol Cell 2018;72:457-468
12. JCI Insight 2019 Sep 5;4(17)
13. Elife 2021 Nov 19;10
14. Cell Rep 2022 Jun 28
15. Cell Rep 2024;43:113753
12/1/2025