Muscle Fibers: Structure & Function
|
Muscle fiber Contraction Crossbridges Excitation-Contraction-Relaxation cycle Structure |
Excitation-Contraction-Relaxation Cycle
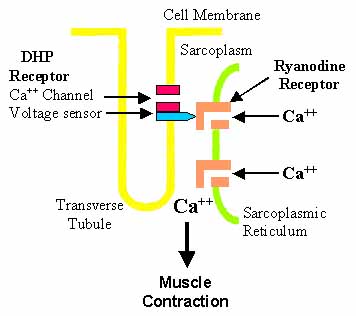
- Motor neuron activity triggers sarcolemmal depolarization via NMJ
- Action potential generated
- Travels along muscle surface membrane
- Enters transverse tubule system (T-tubules)
- Signal transmitted from T-tubules to terminal cisternae at triads
- Ca++ ions released from sarcoplasmic reticulum: Regulated by 2 membrane protein complexes
- Dihydropyridine receptor (DHPR)
- Located in junctional t-tubules
- Voltage sensing: α1 subunit of DHPR activated by membrane depolarization
- Activated DHPR interacts with ryanodyne receptor
- Ryanodyne receptor (RyR)
- Located in junctional sarcoplasmic reticulum (SR)
- RyRs release Ca++ ions into cytosol after interaction with activated DHPR
- Other RyRs in SR membrane without DHPR interactions
- Activated by excess Ca++ ions released by neighboring RyRs
- Amplify & propagate Ca++ signal
- Related molecules: CASQ; Triadin; ATP2A1; STAC3; SPEG
- Dihydropyridine receptor (DHPR)
- Contractile apparatus activated
- Ca++ binds to troponin complex
- Tropomyosin binding to contractile apparatus changes
- Actin allowed to bind to myosin heads
- Muscle contraction occurs via myofilament sliding
Muscle Triads
- Juxtaposition of: T-tubule + 2 Terminal cisternae
- Invaginations of sarcolemma
- 2 Terminal cisternae
- T-tubule
- Structure: External link
- Triad proteins
- Dihydropyridine receptor (DHPR): T-tubule
- Ryanodine receptor (RYR): Sarcoplassmic reticulum
- Triad functions
- Maintain Ca++ homeostasis
- Excitation–contraction (EC) coupling
- EC coupling disorders (Triadopathies)
MUSCLE FIBER STRUCTURE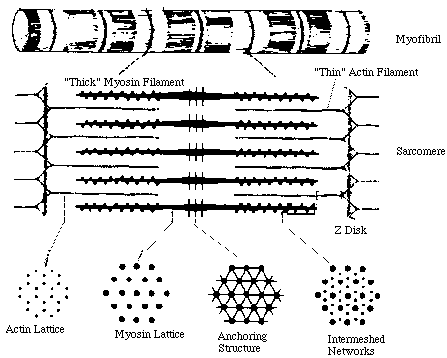 From UCSD THICK-THIN FILAMENT CROSSBRIDGES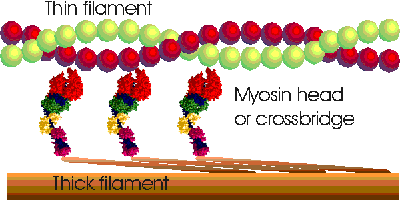
From M Ferenczi MUSCLE FIBER CONTRACTION
From M Ferenczi |
Return to Mysoin & related proteins
Return to Neuromuscular Home Page
4/13/2025